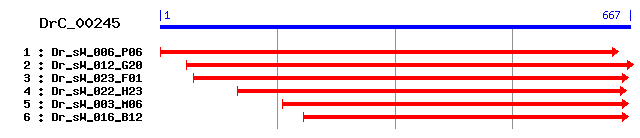
DrC_00245
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00245 (665 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P28797|GRN_CAVPO Granulins precursor (Proepithelin) (PEP... 108 1e-23 sp|P23785|GRN_RAT Granulins precursor [Contains: Acrogranin... 108 1e-23 sp|P28798|GRN_MOUSE Granulins precursor (Acrogranin) (Proep... 106 5e-23 sp|P28799|GRN_HUMAN Granulins precursor (Proepithelin) (PEP... 106 5e-23 sp|P80059|PMD1_LOCMI Pars intercerebralis major peptide D1 ... 63 8e-10 sp|P80930|ENA1_HORSE Antimicrobial peptide eNAP-1 57 4e-08 sp|P81013|GRN1_CYPCA Granulin-1 52 1e-06 sp|P81014|GRN2_CYPCA Granulin-2 49 9e-06 sp|P81015|GRN3_CYPCA Granulin-3 45 2e-04 sp|P43297|RD21A_ARATH Cysteine proteinase RD21a precursor (... 43 8e-04
>sp|P28797|GRN_CAVPO Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7] Length = 591 Score = 108 bits (271), Expect = 1e-23 Identities = 54/157 (34%), Positives = 72/157 (45%) Frame = +1 Query: 73 ITPAFLERSNIINACQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTC 252 + PA L S I C + ++ C D TCC+ SG + CC PNA+CC D +HCCP+ T C Sbjct: 197 LLPAALPTSVI---CPDSRSQCPDDTTCCLLASGEYGCCPMPNAICCSDHLHCCPQDTVC 253 Query: 253 DLVNKRCIHQSNEPIEWPVVKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCP 432 DL RC+ Q+ ++P +T VW+ C CP Sbjct: 254 DLRQSRCLSQNK----------AKTLLTKLPSWT------------VWDVE-CDQEVSCP 290 Query: 433 NSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSGQQC 543 CC L G++ CCPFP VCC C P +C Sbjct: 291 EGQTCCRLQSGKWGCCPFPKAVCCEDHVHCCPERFRC 327
Score = 105 bits (261), Expect = 1e-22
Identities = 56/149 (37%), Positives = 71/149 (47%), Gaps = 6/149 (4%)
Frame = +1
Query: 115 CQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQ-SNE 291
C G+ C D TCC G CC P A CC D++HCCP G +CDLV+ RC+ +
Sbjct: 112 CPGGEFECPDSSTCCHMLDGSWGCCPMPQASCCEDRVHCCPHGASCDLVHIRCVTALGSH 171
Query: 292 PIEWPVVKIVDPAKIEIPKYT-VERNPLRE---FIIKVWNKSTCPDG-SLCPNSFPCCLL 456
P+ + PA + YT E P+ + CPD S CP+ CCLL
Sbjct: 172 PLTTKL-----PA--QRTNYTGAEGTPVVSPGLLPAALPTSVICPDSRSQCPDDTTCCLL 224
Query: 457 APGQYFCCPFPGGVCCSSPPGCVPSGQQC 543
A G+Y CCP P +CCS C P C
Sbjct: 225 ASGEYGCCPMPNAICCSDHLHCCPQDTVC 253
Score = 91.3 bits (225), Expect = 2e-18
Identities = 47/140 (33%), Positives = 60/140 (42%)
Frame = +1
Query: 130 ALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPV 309
A C ++TCC GG CCQ P+AVCC D HCCP G TC++ + C
Sbjct: 445 ASCPVRQTCCPKLGGGWACCQLPHAVCCEDGQHCCPAGYTCNVKARSC------------ 492
Query: 310 VKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFP 489
K D A + P + + C D C + CC + G + CCPF
Sbjct: 493 EKAADGAHLAAPLAVGSTGGVMDV--------ACGDRHFCHDEQTCCRDSRGGWACCPFH 544
Query: 490 GGVCCSSPPGCVPSGQQCLS 549
GVCC C P+G C S
Sbjct: 545 QGVCCKDQRHCCPAGFHCES 564
Score = 73.2 bits (178), Expect = 6e-13
Identities = 42/135 (31%), Positives = 55/135 (40%), Gaps = 6/135 (4%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C + +TCC +SG CC +P AVCC D +HCCPE C C Q + W
Sbjct: 289 CPEGQTCCRLQSGKWGCCPFPKAVCCEDHVHCCPERFRCHTEKDTC-EQGLLQVPWAQKT 347
Query: 316 IVDPAKIEIPK------YTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFC 477
P++ P PLR I +C + C CC LA G++ C
Sbjct: 348 PAQPSRPSQPSPPGPPGPPSPPGPLRSEI-------SCDEVVSCRPGNICCRLASGEWGC 400
Query: 478 CPFPGGVCCSSPPGC 522
CP G C + C
Sbjct: 401 CPSSEGYLCMAGERC 415
Score = 72.4 bits (176), Expect = 1e-12
Identities = 40/144 (27%), Positives = 53/144 (36%), Gaps = 1/144 (0%)
Frame = +1
Query: 115 CQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEP 294
CQ+ A C +C ++ +G CC + A+ CGD HCCP G C CI + +
Sbjct: 47 CQS-PANCPIGHSCVLTAAGTAACCPFSQAMACGDGHHCCPYGFHCSTDGGTCIQRPD-- 103
Query: 295 IEWPVVKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSL-CPNSFPCCLLAPGQY 471
I + CP G CP+S CC + G +
Sbjct: 104 ------------------------------IHLLGAVQCPGGEFECPDSSTCCHMLDGSW 133
Query: 472 FCCPFPGGVCCSSPPGCVPSGQQC 543
CCP P CC C P G C
Sbjct: 134 GCCPMPQASCCEDRVHCCPHGASC 157
Score = 66.2 bits (160), Expect = 7e-11
Identities = 23/62 (37%), Positives = 34/62 (54%)
Frame = +1
Query: 103 IINACQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQ 282
+++ + C D++TCC GG CC + VCC DQ HCCP G C+ RC+H+
Sbjct: 513 VMDVACGDRHFCHDEQTCCRDSRGGWACCPFHQGVCCKDQRHCCPAGFHCESQGTRCVHK 572
Query: 283 SN 288
+
Sbjct: 573 KS 574
Score = 41.2 bits (95), Expect = 0.002
Identities = 16/45 (35%), Positives = 19/45 (42%)
Frame = +1
Query: 409 CPDGSLCPNSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSGQQC 543
C + CP CC G + CC P VCC C P+G C
Sbjct: 441 CDQHASCPVRQTCCPKLGGGWACCQLPHAVCCEDGQHCCPAGYTC 485
>sp|P23785|GRN_RAT Granulins precursor [Contains: Acrogranin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B) (Epithelin-2); Granulin-4 (Granulin A) (Epithelin-1); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)] Length = 588 Score = 108 bits (271), Expect = 1e-23 Identities = 49/143 (34%), Positives = 65/143 (45%) Frame = +1 Query: 115 CQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEP 294 C + K C D TCC +G + CC PNA+CC D +HCCP+ T CDL+ +CI + Sbjct: 206 CPDAKTQCPDDSTCCELPTGKYGCCPMPNAICCSDHLHCCPQDTVCDLIQSKCISKD--- 262 Query: 295 IEWPVVKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYF 474 ++P Y V N+ C CP+ + CC L G + Sbjct: 263 -------YTTDLMTKLPGYPV-------------NEVKCDLEVSCPDGYTCCRLNTGAWG 302 Query: 475 CCPFPGGVCCSSPPGCVPSGQQC 543 CCPF VCC C P+G QC Sbjct: 303 CCPFTKAVCCEDHIHCCPAGFQC 325
Score = 103 bits (258), Expect = 3e-22
Identities = 51/144 (35%), Positives = 66/144 (45%), Gaps = 1/144 (0%)
Frame = +1
Query: 115 CQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEP 294
C + C D TCC+ G CC P A CC D++HCCP G +CDLV+ RCI +
Sbjct: 125 CPGSQFECPDSATCCIMIDGSWGCCPMPQASCCEDRVHCCPHGASCDLVHTRCISPTG-- 182
Query: 295 IEWPVVKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDG-SLCPNSFPCCLLAPGQY 471
P++K K+ +R V CPD + CP+ CC L G+Y
Sbjct: 183 -THPLLK----------KFPAQRTNRAVASFSV----VCPDAKTQCPDDSTCCELPTGKY 227
Query: 472 FCCPFPGGVCCSSPPGCVPSGQQC 543
CCP P +CCS C P C
Sbjct: 228 GCCPMPNAICCSDHLHCCPQDTVC 251
Score = 92.0 bits (227), Expect = 1e-18
Identities = 47/141 (33%), Positives = 64/141 (45%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C D TCC +G CC + AVCC D +HCCP G C C ++ P +K
Sbjct: 287 CPDGYTCCRLNTGAWGCCPFTKAVCCEDHIHCCPAGFQCHTETGTC---ELGVLQVPWMK 343
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFPGG 495
V A + +P + +N + C D S CP++ CC L+ G + CCP P
Sbjct: 344 KVT-ASLSLPDPQILKNDV-----------PCDDFSSCPSNNTCCRLSSGDWGCCPMPEA 391
Query: 496 VCCSSPPGCVPSGQQCLSG*Y 558
VCC C P G +C+ Y
Sbjct: 392 VCCLDHQHCCPQGFKCMDEGY 412
Score = 85.5 bits (210), Expect = 1e-16
Identities = 46/136 (33%), Positives = 60/136 (44%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C +TCC S G CCQ P+AVCC D+ HCCP G TC++ + C
Sbjct: 447 CPVGQTCCPSLKGSWACCQLPHAVCCEDRQHCCPAGYTCNVKARTC-------------- 492
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFPGG 495
E +V+ + F KV N C G C ++ CC + G + CCP+ G
Sbjct: 493 -------EKDAGSVQPSMDLTFGSKVGNVE-CGAGHFCHDNQSCCKDSQGGWACCPYVKG 544
Query: 496 VCCSSPPGCVPSGQQC 543
VCC C P G C
Sbjct: 545 VCCRDGRHCCPIGFHC 560
Score = 79.0 bits (193), Expect = 1e-14
Identities = 39/136 (28%), Positives = 54/136 (39%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C TCC SG CC P AVCC D HCCP+G C +++ + + +
Sbjct: 369 CPSNNTCCRLSSGDWGCCPMPEAVCCLDHQHCCPQGFKC--MDEGYCQKGDRMVAG---- 422
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFPGG 495
+E+ P+R+ + C + CP CC G + CC P
Sbjct: 423 -------------LEKMPVRQTTLLQHGDIGCDQHTSCPVGQTCCPSLKGSWACCQLPHA 469
Query: 496 VCCSSPPGCVPSGQQC 543
VCC C P+G C
Sbjct: 470 VCCEDRQHCCPAGYTC 485
Score = 72.8 bits (177), Expect = 8e-13
Identities = 37/136 (27%), Positives = 49/136 (36%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C D +C ++ SG CC + V C D HCCP G C K C S+ +
Sbjct: 67 CPDGYSCLLTVSGTSSCCPFSEGVSCDDGQHCCPRGFHCSADGKSCSQISDSLL------ 120
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFPGG 495
++ P E CP+S CC++ G + CCP P
Sbjct: 121 ----GAVQCPGSQFE----------------------CPDSATCCIMIDGSWGCCPMPQA 154
Query: 496 VCCSSPPGCVPSGQQC 543
CC C P G C
Sbjct: 155 SCCEDRVHCCPHGASC 170
Score = 58.5 bits (140), Expect = 1e-08
Identities = 24/65 (36%), Positives = 32/65 (49%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C D ++CC GG CC Y VCC D HCCP G C +C+ + W ++
Sbjct: 522 CHDNQSCCKDSQGGWACCPYVKGVCCRDGRHCCPIGFHCSAKGTKCLRKKTP--RWDIL- 578
Query: 316 IVDPA 330
+ DPA
Sbjct: 579 LRDPA 583
Score = 42.4 bits (98), Expect = 0.001
Identities = 17/46 (36%), Positives = 20/46 (43%)
Frame = +1
Query: 406 TCPDGSLCPNSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSGQQC 543
+C CP+ + C L G CCPF GV C C P G C
Sbjct: 60 SCQIRDHCPDGYSCLLTVSGTSSCCPFSEGVSCDDGQHCCPRGFHC 105
>sp|P28798|GRN_MOUSE Granulins precursor (Acrogranin) (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Granulin 1; Granulin 2; Granulin 3; Granulin 4; Granulin 5; Granulin 6; Granulin 7] Length = 589 Score = 106 bits (265), Expect = 5e-23 Identities = 48/143 (33%), Positives = 69/143 (48%) Frame = +1 Query: 115 CQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEP 294 C + K C D TCC +G + CC PNA+CC D +HCCP+ T CDL+ +C+ ++ Sbjct: 207 CPDAKTQCPDDSTCCELPTGKYGCCPMPNAICCSDHLHCCPQDTVCDLIQSKCLSKN--- 263 Query: 295 IEWPVVKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYF 474 ++P Y V+ +K + +CP+G + CC L G + Sbjct: 264 -------YTTDLLTKLPGYPVKE-------VKCDMEVSCPEG------YTCCRLNTGAWG 303 Query: 475 CCPFPGGVCCSSPPGCVPSGQQC 543 CCPF VCC C P+G QC Sbjct: 304 CCPFAKAVCCEDHIHCCPAGFQC 326
Score = 101 bits (252), Expect = 2e-21
Identities = 48/144 (33%), Positives = 62/144 (43%), Gaps = 1/144 (0%)
Frame = +1
Query: 115 CQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEP 294
C + C D TCC+ G CC P A CC D++HCCP G +CDLV+ RC+ +
Sbjct: 125 CPGSQFECPDSATCCIMVDGSWGCCPMPQASCCEDRVHCCPHGASCDLVHTRCVSPTG-- 182
Query: 295 IEWPVVKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDG-SLCPNSFPCCLLAPGQY 471
+ P R F + CPD + CP+ CC L G+Y
Sbjct: 183 --------THTLLKKFPAQKTNRAVSLPFSV------VCPDAKTQCPDDSTCCELPTGKY 228
Query: 472 FCCPFPGGVCCSSPPGCVPSGQQC 543
CCP P +CCS C P C
Sbjct: 229 GCCPMPNAICCSDHLHCCPQDTVC 252
Score = 93.6 bits (231), Expect = 4e-19
Identities = 46/141 (32%), Positives = 61/141 (43%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C + TCC +G CC + AVCC D +HCCP G C C + W + K
Sbjct: 288 CPEGYTCCRLNTGAWGCCPFAKAVCCEDHIHCCPAGFQCHTEKGTC-EMGILQVPW-MKK 345
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFPGG 495
++ P ++ P+ P C D + CP + CC L G + CCP P
Sbjct: 346 VIAPLRLPDPQILKSDTP-------------CDDFTRCPTNNTCCKLNSGDWGCCPIPEA 392
Query: 496 VCCSSPPGCVPSGQQCLSG*Y 558
VCCS C P G CL+ Y
Sbjct: 393 VCCSDNQHCCPQGFTCLAQGY 413
Score = 90.9 bits (224), Expect = 3e-18
Identities = 47/136 (34%), Positives = 64/136 (47%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C +TCC S G CCQ P+AVCC D+ HCCP G TC++ + C + + I+ PV+
Sbjct: 448 CPVGQTCCPSLKGSWACCQLPHAVCCEDRQHCCPAGYTCNVKARTC-EKDVDFIQPPVLL 506
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFPGG 495
+ P KV N C +G C ++ CC + G + CCP+ G
Sbjct: 507 TLGP--------------------KVGNVE-CGEGHFCHDNQTCCKDSAGVWACCPYLKG 545
Query: 496 VCCSSPPGCVPSGQQC 543
VCC C P G C
Sbjct: 546 VCCRDGRHCCPGGFHC 561
Score = 81.3 bits (199), Expect = 2e-15
Identities = 47/136 (34%), Positives = 57/136 (41%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C TCC SG CC P AVCC D HCCP+G TC L C Q + + + K
Sbjct: 370 CPTNNTCCKLNSGDWGCCPIPEAVCCSDNQHCCPQGFTC-LAQGYC--QKGDTMVAGLEK 426
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFPGG 495
I PA+ + PL+ I C + CP CC G + CC P
Sbjct: 427 I--PAR--------QTTPLQIGDI------GCDQHTSCPVGQTCCPSLKGSWACCQLPHA 470
Query: 496 VCCSSPPGCVPSGQQC 543
VCC C P+G C
Sbjct: 471 VCCEDRQHCCPAGYTC 486
Score = 79.0 bits (193), Expect = 1e-14
Identities = 38/136 (27%), Positives = 51/136 (37%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C +C ++ SG CC + V CGD HCCP+G C K C S+ P+
Sbjct: 67 CPAGYSCLLTVSGTSSCCPFSKGVSCGDGYHCCPQGFHCSADGKSCFQMSDNPL------ 120
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFPGG 495
++ P E CP+S CC++ G + CCP P
Sbjct: 121 ----GAVQCPGSQFE----------------------CPDSATCCIMVDGSWGCCPMPQA 154
Query: 496 VCCSSPPGCVPSGQQC 543
CC C P G C
Sbjct: 155 SCCEDRVHCCPHGASC 170
Score = 55.1 bits (131), Expect = 2e-07
Identities = 20/49 (40%), Positives = 25/49 (51%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQ 282
C D +TCC +G CC Y VCC D HCCP G C +C+ +
Sbjct: 523 CHDNQTCCKDSAGVWACCPYLKGVCCRDGRHCCPGGFHCSARGTKCLRK 571
Score = 41.6 bits (96), Expect = 0.002
Identities = 17/46 (36%), Positives = 19/46 (41%)
Frame = +1
Query: 406 TCPDGSLCPNSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSGQQC 543
+C CP + C L G CCPF GV C C P G C
Sbjct: 60 SCQTHGHCPAGYSCLLTVSGTSSCCPFSKGVSCGDGYHCCPQGFHC 105
>sp|P28799|GRN_HUMAN Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)] Length = 593 Score = 106 bits (265), Expect = 5e-23 Identities = 49/143 (34%), Positives = 69/143 (48%) Frame = +1 Query: 115 CQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEP 294 C + ++ C D TCC SG + CC PNA CC D +HCCP+ T CDL+ +C+ + N Sbjct: 208 CPDARSRCPDGSTCCELPSGKYGCCPMPNATCCSDHLHCCPQDTVCDLIQSKCLSKENAT 267 Query: 295 IEWPVVKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYF 474 + ++P +TV +K + +CPDG + CC L G + Sbjct: 268 TD---------LLTKLPAHTVGD-------VKCDMEVSCPDG------YTCCRLQSGAWG 305 Query: 475 CCPFPGGVCCSSPPGCVPSGQQC 543 CCPF VCC C P+G C Sbjct: 306 CCPFTQAVCCEDHIHCCPAGFTC 328
Score = 104 bits (259), Expect = 2e-22
Identities = 51/144 (35%), Positives = 64/144 (44%), Gaps = 1/144 (0%)
Frame = +1
Query: 115 CQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEP 294
C + + C D TCCV G CC P A CC D++HCCP G CDLV+ RCI +
Sbjct: 126 CPDSQFECPDFSTCCVMVDGSWGCCPMPQASCCEDRVHCCPHGAFCDLVHTRCITPTG-- 183
Query: 295 IEWPVVKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDG-SLCPNSFPCCLLAPGQY 471
P ++P R + + + CPD S CP+ CC L G+Y
Sbjct: 184 --------THPLAKKLPAQRTNR------AVALSSSVMCPDARSRCPDGSTCCELPSGKY 229
Query: 472 FCCPFPGGVCCSSPPGCVPSGQQC 543
CCP P CCS C P C
Sbjct: 230 GCCPMPNATCCSDHLHCCPQDTVC 253
Score = 100 bits (249), Expect = 3e-21
Identities = 49/138 (35%), Positives = 63/138 (45%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C D TCC +SG CC + AVCC D +HCCP G TCD C Q + W
Sbjct: 290 CPDGYTCCRLQSGAWGCCPFTQAVCCEDHIHCCPAGFTCDTQKGTC-EQGPHQVPWMEKA 348
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYFCCPFPGG 495
PA + +P + + C + S CP+S CC L G++ CCP P
Sbjct: 349 ---PAHLSLPDPQALKRDV-----------PCDNVSSCPSSDTCCQLTSGEWGCCPIPEA 394
Query: 496 VCCSSPPGCVPSGQQCLS 549
VCCS C P G C++
Sbjct: 395 VCCSDHQHCCPQGYTCVA 412
Score = 83.6 bits (205), Expect = 4e-16
Identities = 47/137 (34%), Positives = 63/137 (45%), Gaps = 1/137 (0%)
Frame = +1
Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVK 315
C +TCC S G CCQ P+AVCC D+ HCCP G TC++ + C E VV
Sbjct: 450 CPVGQTCCPSLGGSWACCQLPHAVCCEDRQHCCPAGYTCNVKARSC--------EKEVVS 501
Query: 316 IVDPAKIEIPKYTVERNPLREFIIKVWNKST-CPDGSLCPNSFPCCLLAPGQYFCCPFPG 492
P + R+P V K C +G C ++ CC + CCP+
Sbjct: 502 AQ-------PATFLARSP------HVGVKDVECGEGHFCHDNQTCCRDNRQGWACCPYRQ 548
Query: 493 GVCCSSPPGCVPSGQQC 543
GVCC+ C P+G +C
Sbjct: 549 GVCCADRRHCCPAGFRC 565
Score = 81.3 bits (199), Expect = 2e-15
Identities = 46/143 (32%), Positives = 57/143 (39%)
Frame = +1
Query: 115 CQNGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEP 294
C N + C TCC SG CC P AVCC D HCCP+G TC + +C S
Sbjct: 366 CDNVSS-CPSSDTCCQLTSGEWGCCPIPEAVCCSDHQHCCPQGYTC-VAEGQCQRGS--- 420
Query: 295 IEWPVVKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSLCPNSFPCCLLAPGQYF 474
+IV +E+ P R + C + CP CC G +
Sbjct: 421 ------EIVAG---------LEKMPARRASLSHPRDIGCDQHTSCPVGQTCCPSLGGSWA 465
Query: 475 CCPFPGGVCCSSPPGCVPSGQQC 543
CC P VCC C P+G C
Sbjct: 466 CCQLPHAVCCEDRQHCCPAGYTC 488
Score = 77.0 bits (188), Expect = 4e-14
Identities = 39/139 (28%), Positives = 49/139 (35%), Gaps = 1/139 (0%)
Frame = +1
Query: 130 ALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPV 309
A C +C + SG CC +P AV CGD HCCP G C + C +S +
Sbjct: 65 AHCSAGHSCIFTVSGTSSCCPFPEAVACGDGHHCCPRGFHCSADGRSCFQRSGNNSVGAI 124
Query: 310 VKIVDPAKIEIPKYTVERNPLREFIIKVWNKSTCPDGSL-CPNSFPCCLLAPGQYFCCPF 486
CPD CP+ CC++ G + CCP
Sbjct: 125 --------------------------------QCPDSQFECPDFSTCCVMVDGSWGCCPM 152
Query: 487 PGGVCCSSPPGCVPSGQQC 543
P CC C P G C
Sbjct: 153 PQASCCEDRVHCCPHGAFC 171
Score = 62.8 bits (151), Expect = 8e-10
Identities = 37/112 (33%), Positives = 49/112 (43%), Gaps = 5/112 (4%)
Frame = +1
Query: 10 CCVSTQRFSIVTSILVQYIVSITPA-FLERSNIINA----CQNGKALCGDQKTCCVSKSG 174
CC + ++ + +VS PA FL RS + C G C D +TCC
Sbjct: 481 CCPAGYTCNVKARSCEKEVVSAQPATFLARSPHVGVKDVECGEGH-FCHDNQTCCRDNRQ 539
Query: 175 GHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCIHQSNEPIEWPVVKIVDPA 330
G CC Y VCC D+ HCCP G C +C+ + E W + DPA
Sbjct: 540 GWACCPYRQGVCCADRRHCCPAGFRCAARGTKCLRR--EAPRWD-APLRDPA 588
Score = 36.2 bits (82), Expect = 0.079
Identities = 15/45 (33%), Positives = 16/45 (35%)
Frame = +1
Query: 409 CPDGSLCPNSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSGQQC 543
C + C C G CCPFP V C C P G C
Sbjct: 61 CQVDAHCSAGHSCIFTVSGTSSCCPFPEAVACGDGHHCCPRGFHC 105
>sp|P80059|PMD1_LOCMI Pars intercerebralis major peptide D1 (PMP-D1) Length = 54 Score = 62.8 bits (151), Expect = 8e-10 Identities = 23/47 (48%), Positives = 28/47 (59%) Frame = +1 Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCI 276 C +TCC + G CC Y VCC D +HCCP GT CD ++RCI Sbjct: 7 CPGTETCCTTPQGEEGCCPYKEGVCCLDGIHCCPSGTVCDEDHRRCI 53
Score = 45.1 bits (105), Expect = 2e-04
Identities = 17/39 (43%), Positives = 20/39 (51%)
Frame = +1
Query: 427 CPNSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSGQQC 543
CP + CC G+ CCP+ GVCC C PSG C
Sbjct: 7 CPGTETCCTTPQGEEGCCPYKEGVCCLDGIHCCPSGTVC 45
>sp|P80930|ENA1_HORSE Antimicrobial peptide eNAP-1 Length = 46 Score = 57.0 bits (136), Expect = 4e-08 Identities = 20/36 (55%), Positives = 22/36 (61%) Frame = +1 Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEG 243 C D +TCC + GG CC Y VCC DQ HCCP G Sbjct: 10 CHDXQTCCRASQGGXACCPYSQGVCCADQRHCCPVG 45
Score = 43.5 bits (101), Expect = 5e-04
Identities = 16/42 (38%), Positives = 21/42 (50%)
Frame = +1
Query: 409 CPDGSLCPNSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSG 534
C +G C + CC + G CCP+ GVCC+ C P G
Sbjct: 4 CGEGHFCHDXQTCCRASQGGXACCPYSQGVCCADQRHCCPVG 45
>sp|P81013|GRN1_CYPCA Granulin-1 Length = 57 Score = 52.0 bits (123), Expect = 1e-06 Identities = 19/48 (39%), Positives = 25/48 (52%) Frame = +1 Query: 133 LCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCI 276 +C D TCC+S G CC + CC D +HCC G CD + C+ Sbjct: 9 ICPDGTTCCLSPYGVWYCCPFSMGQCCRDGIHCCRHGYHCDSTSTHCL 56
Score = 46.2 bits (108), Expect = 8e-05
Identities = 17/45 (37%), Positives = 22/45 (48%)
Frame = +1
Query: 409 CPDGSLCPNSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSGQQC 543
C ++CP+ CCL G ++CCPF G CC C G C
Sbjct: 4 CDAATICPDGTTCCLSPYGVWYCCPFSMGQCCRDGIHCCRHGYHC 48
>sp|P81014|GRN2_CYPCA Granulin-2 Length = 57 Score = 49.3 bits (116), Expect = 9e-06 Identities = 19/52 (36%), Positives = 24/52 (46%) Frame = +1 Query: 121 NGKALCGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCI 276 N + C + TCC S G CC + CC D HCC G CD + C+ Sbjct: 5 NARTTCPSRTTCCRSPFGVWYCCPFLMGQCCRDGRHCCRHGYRCDSTSTLCL 56
Score = 40.8 bits (94), Expect = 0.003
Identities = 16/45 (35%), Positives = 21/45 (46%)
Frame = +1
Query: 409 CPDGSLCPNSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSGQQC 543
C + CP+ CC G ++CCPF G CC C G +C
Sbjct: 4 CNARTTCPSRTTCCRSPFGVWYCCPFLMGQCCRDGRHCCRHGYRC 48
>sp|P81015|GRN3_CYPCA Granulin-3 Length = 57 Score = 45.1 bits (105), Expect = 2e-04 Identities = 18/47 (38%), Positives = 21/47 (44%) Frame = +1 Query: 136 CGDQKTCCVSKSGGHQCCQYPNAVCCGDQMHCCPEGTTCDLVNKRCI 276 C TCC S G CC + CC D HCC G CD + C+ Sbjct: 10 CPSGTTCCRSPFGVWYCCPFLMGQCCRDGRHCCRHGYHCDSTSTLCL 56
Score = 43.1 bits (100), Expect = 6e-04
Identities = 17/45 (37%), Positives = 20/45 (44%)
Frame = +1
Query: 409 CPDGSLCPNSFPCCLLAPGQYFCCPFPGGVCCSSPPGCVPSGQQC 543
C G CP+ CC G ++CCPF G CC C G C
Sbjct: 4 CDAGITCPSGTTCCRSPFGVWYCCPFLMGQCCRDGRHCCRHGYHC 48
>sp|P43297|RD21A_ARATH Cysteine proteinase RD21a precursor (RD21) Length = 462 Score = 42.7 bits (99), Expect = 8e-04 Identities = 22/66 (33%), Positives = 25/66 (37%), Gaps = 6/66 (9%) Frame = +1 Query: 136 CGDQKTCCVSKSGGHQC-----CQYPNAVCCGDQMHCCP-EGTTCDLVNKRCIHQSNEPI 297 C + TCC G C C A CC D CCP E CDL C+ N P Sbjct: 381 CPESNTCCCLFEYGKYCFAWGCCPLEAATCCDDNYSCCPHEYPVCDLDQGTCLLSKNSPF 440 Query: 298 EWPVVK 315 +K Sbjct: 441 SVKALK 446
Score = 36.6 bits (83), Expect = 0.061
Identities = 16/45 (35%), Positives = 19/45 (42%), Gaps = 4/45 (8%)
Frame = +1
Query: 406 TCPDGSLCPNSFPCCLLAPGQYF----CCPFPGGVCCSSPPGCVP 528
TCP+ + C CCL G+Y CCP CC C P
Sbjct: 380 TCPESNTC-----CCLFEYGKYCFAWGCCPLEAATCCDDNYSCCP 419
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 82,260,404
Number of Sequences: 369166
Number of extensions: 1948029
Number of successful extensions: 6279
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5310
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6136
length of database: 68,354,980
effective HSP length: 106
effective length of database: 48,773,070
effective search space used: 5608903050
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail