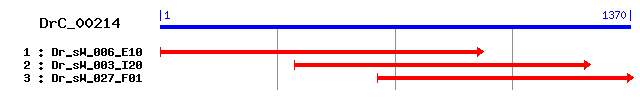
DrC_00214
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00214 (1370 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P97329|KI20A_MOUSE Kinesin family member 20A (Rabkinesin... 39 0.035 sp|O95235|KI20A_HUMAN Kinesin family member 20A (Rabkinesin... 37 0.10 sp|Q06106|MRD1_YEAST Multiple RNA-binding domain-containing... 37 0.13 sp|Q9JMH9|MY18A_MOUSE Myosin-18A (Myosin XVIIIa) (Myosin co... 34 0.87 sp|P08799|MYS2_DICDI Myosin II heavy chain, non muscle 34 0.87 sp|P25386|USO1_YEAST Intracellular protein transport protei... 34 1.1 sp|O67825|IF2_AQUAE Translation initiation factor IF-2 34 1.1 sp|Q07847|VE1_HPV63 Replication protein E1 33 1.5 sp|P02566|MYO4_CAEEL Myosin-4 (Myosin heavy chain B) (MHC B... 32 3.3 sp|P36353|VNS5_MSTV Nonstructural protein NS5 32 3.3
>sp|P97329|KI20A_MOUSE Kinesin family member 20A (Rabkinesin-6) (Rab6-interacting kinesin-like protein) (Kinesin-like protein 174) Length = 887 Score = 38.9 bits (89), Expect = 0.035 Identities = 26/107 (24%), Positives = 48/107 (44%), Gaps = 3/107 (2%) Frame = +1 Query: 301 QEIEEFDDEYVPLPDNLPERSKE---EMETLLWTQKRDSQIREWQEVKQLEREKVKLEHQ 471 ++I+E D++ L L E ++ + L +R ++ +Q + K +LE Sbjct: 638 EQIQERDEKIEELETLLQEAKQQPAAQQSGGLSLLRRSQRLAASASTQQFQEVKAELEQC 697 Query: 472 VRSMQRRNSRAHKTSDILKVPPFAKPHHLAIDQDSEDKRFQFPLLTT 612 + + HK +LK PP AKP + +D+ E+ + LL T Sbjct: 698 KTELSSTTAELHKYQQVLKPPPPAKPFTIDVDKKLEEGQKNIRLLRT 744
>sp|O95235|KI20A_HUMAN Kinesin family member 20A (Rabkinesin-6) (Rab6-interacting kinesin-like protein) (GG10_2) Length = 890 Score = 37.4 bits (85), Expect = 0.10 Identities = 28/111 (25%), Positives = 49/111 (44%), Gaps = 7/111 (6%) Frame = +1 Query: 301 QEIEEFDDEYVPLPDNLPERSKEEM-------ETLLWTQKRDSQIREWQEVKQLEREKVK 459 +EI+E D++ L L E ++ + E L +R ++ +QL+ K K Sbjct: 639 EEIQERDEKIEELEALLQEARQQSVAHQQSGSELAL---RRSQRLAASASTQQLQEVKAK 695 Query: 460 LEHQVRSMQRRNSRAHKTSDILKVPPFAKPHHLAIDQDSEDKRFQFPLLTT 612 L+ + HK +L+ PP AKP + +D+ E+ + LL T Sbjct: 696 LQQCKAELNSTTEELHKYQKMLEPPPSAKPFTIDVDKKLEEGQKNIRLLRT 746
>sp|Q06106|MRD1_YEAST Multiple RNA-binding domain-containing protein 1 Length = 887 Score = 37.0 bits (84), Expect = 0.13 Identities = 25/92 (27%), Positives = 48/92 (52%), Gaps = 3/92 (3%) Frame = +1 Query: 358 RSKEEMETLLWTQKRDSQIREWQEV---KQLEREKVKLEHQVRSMQRRNSRAHKTSDILK 528 R ++++ + T K SQ+ W++V K +E EK+K E + S+Q + AH + + + Sbjct: 146 RKNKQLQEFMETMKPSSQVTSWEKVGIDKSIEDEKLKREEEDSSVQGNSLLAHALA-LKE 204 Query: 529 VPPFAKPHHLAIDQDSEDKRFQFPLLTTNDDE 624 + +L I+ +S+D ++ L N DE Sbjct: 205 ENNKDEAPNLVIENESDD---EYSALNRNRDE 233
>sp|Q9JMH9|MY18A_MOUSE Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN) Length = 2035 Score = 34.3 bits (77), Expect = 0.87 Identities = 28/100 (28%), Positives = 51/100 (51%), Gaps = 7/100 (7%) Frame = +1 Query: 298 LQEIE---EFDDEYVPLPDNLPERSKEEMETLLWTQKRD----SQIREWQEVKQLEREKV 456 ++E+E EF+ V +NL R KE ME L T++RD ++ RE ++ K+L+R+ Sbjct: 1820 IRELETRLEFEKTQVKRLENLASRLKETMEKL--TEERDQRAAAENREKEQNKRLQRQLR 1877 Query: 457 KLEHQVRSMQRRNSRAHKTSDILKVPPFAKPHHLAIDQDS 576 + ++ + R+ + A + K H L +D +S Sbjct: 1878 DTKEEMSELARKEAEASR-----------KKHELEMDLES 1906
>sp|P08799|MYS2_DICDI Myosin II heavy chain, non muscle Length = 2116 Score = 34.3 bits (77), Expect = 0.87 Identities = 37/152 (24%), Positives = 72/152 (47%), Gaps = 8/152 (5%) Frame = +1 Query: 253 SSSIAKMAGIQNPDLLQEIEEFDDEYVPLPDNLPERSK-----EEMETLLWTQKRDSQIR 417 S +I+++ I++ +L +E+EE L ++ E SK E+ L ++ D +R Sbjct: 967 SDTISRLEKIKD-ELQKEVEE-------LTESFSEESKDKGVLEKTRVRLQSELDDLTVR 1018 Query: 418 ---EWQEVKQLEREKVKLEHQVRSMQRRNSRAHKTSDILKVPPFAKPHHLAIDQDSEDKR 588 E ++ +L R+K KLE +++ +Q A K+ A L + +++ Sbjct: 1019 LDSETKDKSELLRQKKKLEEELKQVQ----EALAAETAAKLAQEAANKKLQGEYTELNEK 1074 Query: 589 FQFPLLTTNDDEKTMKTFNVQLSNFNDKLMEQ 684 F + ++ EK+ KT QL N++L E+ Sbjct: 1075 FNSEVTARSNVEKSKKTLESQLVAVNNELDEE 1106
>sp|P25386|USO1_YEAST Intracellular protein transport protein USO1 (Int-1) Length = 1790 Score = 33.9 bits (76), Expect = 1.1 Identities = 35/187 (18%), Positives = 83/187 (44%), Gaps = 3/187 (1%) Frame = +1 Query: 169 DKSECIYDSKVKLSANCDLLIQNDWTSNSSSIAKMAGIQNPDLLQEIEEFDDE---YVPL 339 D S+ Y+S++ L ++ T+N ++ K++ +L + EE + E Y L Sbjct: 1046 DSSKDEYESQISLLKE---KLETATTANDENVNKIS-----ELTKTREELEAELAAYKNL 1097 Query: 340 PDNLPERSKEEMETLLWTQKRDSQIREWQEVKQLEREKVKLEHQVRSMQRRNSRAHKTSD 519 + L + + + L ++ + ++E E QLE+E + + Q+ S++ K + Sbjct: 1098 KNELETKLETSEKALKEVKENEEHLKE--EKIQLEKEATETKQQLNSLRANLESLEKEHE 1155 Query: 520 ILKVPPFAKPHHLAIDQDSEDKRFQFPLLTTNDDEKTMKTFNVQLSNFNDKLMEQINIVK 699 L +A ++++++ + ND+ + + N + ND+L ++ +K Sbjct: 1156 DLAAQLKKYEEQIA----NKERQYNEEISQLNDEITSTQQENESIKKKNDELEGEVKAMK 1211 Query: 700 NNKTTKS 720 + +S Sbjct: 1212 STSEEQS 1218
>sp|O67825|IF2_AQUAE Translation initiation factor IF-2 Length = 805 Score = 33.9 bits (76), Expect = 1.1 Identities = 33/159 (20%), Positives = 72/159 (45%), Gaps = 1/159 (0%) Frame = +1 Query: 244 TSNSSSIAKMAGIQNPDLLQEIEEFDDEYVPLPDNLPERSKEEMETLLWTQKRDS-QIRE 420 T S +AK G+++ +EI EF +EY P PD P ++ ++ D+ I+E Sbjct: 6 TKRVSDVAKELGVKS----KEIIEFLNEYYPRPDGKPWKASHGLDEQALEMIYDAFGIKE 61 Query: 421 WQEVKQLEREKVKLEHQVRSMQRRNSRAHKTSDILKVPPFAKPHHLAIDQDSEDKRFQFP 600 +E +++ E+ + +V +++ + + +V KP I ++ E+K+ + Sbjct: 62 EEEKEEVVTEQAQAPAEVE--EKKEEEKKEEVIVEEVVEEKKPE--VIVEEIEEKKEEEE 117 Query: 601 LLTTNDDEKTMKTFNVQLSNFNDKLMEQINIVKNNKTTK 717 +K+++ ++ +K E+ + K K K Sbjct: 118 KKEEEKPKKSVEELIKEILEKKEKEKEKKKVEKERKEEK 156
>sp|Q07847|VE1_HPV63 Replication protein E1 Length = 618 Score = 33.5 bits (75), Expect = 1.5 Identities = 31/143 (21%), Positives = 57/143 (39%), Gaps = 10/143 (6%) Frame = +1 Query: 304 EIEEFDDEYVPLPDNLPERSKEEMETLLWTQKRDSQIREWQEVKQLEREKVKLEHQVRSM 483 E+E+ +D Y ++L RS+ ++ LL + +Q + ++L H + Sbjct: 26 ELEDLEDTY----NSLFNRSESDISDLL------------DDTQQSQGNSLELFHLQEHL 69 Query: 484 QRRNSRAHKTSDILKVPPFAK----------PHHLAIDQDSEDKRFQFPLLTTNDDEKTM 633 Q L PP A P +I +K+ + L T ND + Sbjct: 70 QNEQDLNTLKRKYLNSPPQASATETACNSLSPRLESITISQREKKARKQLFTQNDSGIEL 129 Query: 634 KTFNVQLSNFNDKLMEQINIVKN 702 ++ N N+ L EQ++IV++ Sbjct: 130 SLCQDEVDNINEALQEQVDIVQS 152
>sp|P02566|MYO4_CAEEL Myosin-4 (Myosin heavy chain B) (MHC B) (Uncoordinated protein 54) Length = 1966 Score = 32.3 bits (72), Expect = 3.3 Identities = 26/110 (23%), Positives = 54/110 (49%), Gaps = 4/110 (3%) Frame = +1 Query: 169 DKSECIYDSKVKLSANCDLLIQNDWTSNSSSIAKMAGI--QNPDLLQEIEEFDDEYVPLP 342 + S + + K L N +++ T S + ++A + Q D +++ E +D+ Sbjct: 887 ESSAKLVEEKTSLFTN----LESTKTQLSDAEERLAKLEAQQKDASKQLSELNDQLADNE 942 Query: 343 DNLPE--RSKEEMETLLWTQKRDSQIREWQEVKQLEREKVKLEHQVRSMQ 486 D + R+K+++E + K+ Q E +++ E EK +HQ+RS+Q Sbjct: 943 DRTADVQRAKKKIEAEVEALKKQIQDLE-MSLRKAESEKQSKDHQIRSLQ 991
>sp|P36353|VNS5_MSTV Nonstructural protein NS5 Length = 375 Score = 32.3 bits (72), Expect = 3.3 Identities = 28/108 (25%), Positives = 51/108 (47%), Gaps = 3/108 (2%) Frame = +1 Query: 151 DCHFDFDKSECIYDSKVKLSANCDLLIQNDWTSNSSSIAKMAGIQNP--DLLQEIEEFDD 324 D F + K C +++ + C DW S S+ +Q+P DL+ Sbjct: 242 DQEFRYWKGHCSFET-TRPRVRC-----GDWQSPLDSLV---ALQSPPMDLVCNAPTLLP 292 Query: 325 EYVPLPDNLPERSKEEMETLLWTQKRDSQIREWQE-VKQLEREKVKLE 465 +P+PD+ P K ++ L WT++ + +RE Q+ +K+ EK K++ Sbjct: 293 TAIPVPDH-PSYRKYKLSDLSWTKQHERAVREDQKRLKRAVGEKDKIK 339
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 139,704,330
Number of Sequences: 369166
Number of extensions: 2772247
Number of successful extensions: 9197
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8593
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9177
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16174954980
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail