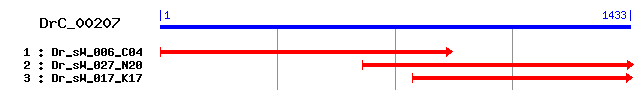
DrC_00207
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00207 (1433 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9Y6P5|SESN1_HUMAN Sestrin-1 (p53-regulated protein PA26) 130 1e-29 sp|Q9CYP7|SESN3_MOUSE Sestrin-3 127 1e-28 sp|P58006|SESN1_MOUSE Sestrin-1 (p53-regulated protein PA26) 125 2e-28 sp|P58005|SESN3_HUMAN Sestrin-3 124 5e-28 sp|P58004|SESN2_HUMAN Sestrin-2 (Hi95) 124 7e-28 sp|P58043|SESN2_MOUSE Sestrin-2 121 4e-27 sp|P58003|SESN1_XENLA Sestrin-1 (p53-regulated protein PA26... 117 8e-26 sp|Q9W1K5|SEST_DROME Putative sestrin 115 4e-25 sp|Q9N4D6|SEST_CAEEL Putative sestrin 74 8e-13 sp|Q9Y7J8|YGL1_SCHPO Hypothetical protein C216.01c in chrom... 38 0.064
>sp|Q9Y6P5|SESN1_HUMAN Sestrin-1 (p53-regulated protein PA26) Length = 492 Score = 130 bits (326), Expect = 1e-29 Identities = 88/313 (28%), Positives = 149/313 (47%), Gaps = 14/313 (4%) Frame = +3 Query: 15 RFYLAIMAAAQMHCKELIYRFITEFIAAGGDPNWLQGLHKAPSRFRPLAVLNQHLCHQPW 194 R Y+ IMAAA+ C L+ + +F+ GGDP WL GL AP + + L LN+ L H+PW Sbjct: 117 RHYIGIMAAARHQCSYLVNLHVNDFLHVGGDPKWLNGLENAPQKLQNLGELNKVLAHRPW 176 Query: 195 QIEPKNIQQLIHSPKGLFHISMSQLMQAVVIMTHVYSLSSFILVCGINSEID-------H 353 I ++I+ L+ + + + S+++L+ AVV++TH +SL+SF CGI+ EI Sbjct: 177 LITKEHIEGLLKAEEHSW--SLAELVHAVVLLTHYHSLASFTFGCGISPEIHCDGGHTFR 234 Query: 354 PPALLWY---NSTNSDSSNSLRNTHSTENISLSNSNDVXXXXXXXXXXXXXXXXXXXXQS 524 PP++ Y + TN + S +S EN+S+S+S Sbjct: 235 PPSVSNYCICDITNGNHSVDEMPVNSAENVSVSDS------------------------- 269 Query: 525 ETDDDKSDENHEDYPLDVLMERLRLAQXXXXXXXXXXXXKMFLSVEKENETIIKNQNHPK 704 + ++ LME++R Q ++ E++ + + Sbjct: 270 ------------FFEVEALMEKMRQLQECRDEEEASQEEMASRFEIEKRESMFVFSSDDE 317 Query: 705 EVNP--LIERFIEDPNFTYISYNSNNAEV--LRMQEYSWSDEACCLADRFYTDIGKALED 872 EV P + R ED ++ Y ++ + V R+Q+Y W D L +R Y D+G+ +++ Sbjct: 318 EVTPARAVSRHFEDTSYGYKDFSRHGMHVPTFRVQDYCWEDHGYSLVNRLYPDVGQLIDE 377 Query: 873 KFNCIIGKNI*YN 911 KF+ I N+ YN Sbjct: 378 KFH--IAYNLTYN 388
Score = 87.0 bits (214), Expect = 1e-16
Identities = 40/102 (39%), Positives = 66/102 (64%)
Frame = +1
Query: 898 TYNTMGDKTNIDTTIFRLSVWIYVHSLYGIRHDDFNYAQMDILLTNPLKTFLKKAMTKPC 1077
TYNTM ++DT++ R ++W Y+H ++GIR+DD++Y +++ LL K ++K + P
Sbjct: 386 TYNTMAMHKDVDTSMLRRAIWNYIHCMFGIRYDDYDYGEINQLLDRSFKVYIKTVVCTPE 445
Query: 1078 TSKKSDLFRFFPEFKISEKVHIALMAADARQQACLLHILKAM 1203
K F+ +FK SEKVH+ L+ +AR QA LL+ L+A+
Sbjct: 446 KVTKRMYDSFWRQFKHSEKVHVNLLLIEARMQAELLYALRAI 487
>sp|Q9CYP7|SESN3_MOUSE Sestrin-3 Length = 492 Score = 127 bits (318), Expect = 1e-28 Identities = 93/308 (30%), Positives = 143/308 (46%), Gaps = 5/308 (1%) Frame = +3 Query: 3 PYVDRFYLAIMAAAQMHCKELIYRFITEFIAAGGDPNWLQGLHKAPSRFRPLAVLNQHLC 182 P DR Y+AIMAAA+ C LI + EF+ GG WL GL P R R L +N+ L Sbjct: 104 PLPDRHYIAIMAAARHQCSYLINMHVDEFLKTGGIAEWLNGLEYVPQRLRNLNEINKLLA 163 Query: 183 HQPWQIEPKNIQQLIHSPKGLFHISMSQLMQAVVIMTHVYSLSSFILVCGINSEIDHPPA 362 H+PW I ++IQ+L+ + G + S+ +L+ AVV++ H ++L+SF+ GIN E D P Sbjct: 164 HRPWLITKEHIQKLVKT--GENNWSLPELVHAVVLLAHYHALASFVFGSGINPERD-PGI 220 Query: 363 LLWYN--STNSDSSNSLRNTHSTENISLSNSNDVXXXXXXXXXXXXXXXXXXXXQSETDD 536 + S +S L N +S EN SL+ SN Q + +D Sbjct: 221 ANGFRLISVSSFCVCDLANDNSIENTSLAGSN--FGIVDSLGELEALMERMKRLQEDRED 278 Query: 537 DKSDENHEDYPLDVLMERLRLAQXXXXXXXXXXXXKMFLSVEKENETIIKNQNHPKEVNP 716 D++ + + R + F + E++ I+ Sbjct: 279 DETTR-------EEMTTRFEKEKKESLFVVPGETLHAFPHSDFEDDVIVTAD-------- 323 Query: 717 LIERFIEDPNFTYISYNSNNAEVL---RMQEYSWSDEACCLADRFYTDIGKALEDKFNCI 887 + R+IEDP+F Y + E L R Q+Y+W + L +R Y+DIG L++KF + Sbjct: 324 -VSRYIEDPSFGYEDFARRGEEHLPTFRAQDYTWENHGFSLVNRLYSDIGHLLDEKFRMV 382 Query: 888 IGKNI*YN 911 N+ YN Sbjct: 383 Y--NLTYN 388
Score = 82.4 bits (202), Expect = 3e-15
Identities = 40/102 (39%), Positives = 65/102 (63%)
Frame = +1
Query: 898 TYNTMGDKTNIDTTIFRLSVWIYVHSLYGIRHDDFNYAQMDILLTNPLKTFLKKAMTKPC 1077
TYNTM ++DTT R +++ YVH ++GIR+DD++Y +++ LL LK ++K P
Sbjct: 386 TYNTMATHEDVDTTTLRRALFNYVHCMFGIRYDDYDYGEVNQLLERSLKVYIKTVTCYPE 445
Query: 1078 TSKKSDLFRFFPEFKISEKVHIALMAADARQQACLLHILKAM 1203
+ K ++ +F SEKVH+ L+ +AR QA LL+ L+A+
Sbjct: 446 RTTKRMYDSYWRQFTHSEKVHVNLLLMEARMQAELLYALRAI 487
>sp|P58006|SESN1_MOUSE Sestrin-1 (p53-regulated protein PA26) Length = 492 Score = 125 bits (315), Expect = 2e-28 Identities = 87/313 (27%), Positives = 147/313 (46%), Gaps = 14/313 (4%) Frame = +3 Query: 15 RFYLAIMAAAQMHCKELIYRFITEFIAAGGDPNWLQGLHKAPSRFRPLAVLNQHLCHQPW 194 R Y+ IMAAA+ C L+ +++F+ GGDP WL GL AP + + L LN+ L H+PW Sbjct: 117 RHYIGIMAAARHQCSYLVNLHVSDFLHVGGDPKWLNGLENAPQKLQNLGELNKVLAHRPW 176 Query: 195 QIEPKNIQQLIHSPKGLFHISMSQLMQAVVIMTHVYSLSSFILVCGINSEID-------H 353 I ++I+ L+ + + + S+++L+ AVV++TH +SL+SF CGI+ EI Sbjct: 177 LITKEHIEGLLKAEEHSW--SLAELVHAVVLLTHYHSLASFTFGCGISPEIHCDGGHTFR 234 Query: 354 PPALLWY---NSTNSDSSNSLRNTHSTENISLSNSNDVXXXXXXXXXXXXXXXXXXXXQS 524 PP++ Y + TN + S +S N S+S+S Sbjct: 235 PPSVSNYCICDITNGNHSVDEMQVNSAGNASVSDS------------------------- 269 Query: 525 ETDDDKSDENHEDYPLDVLMERLRLAQXXXXXXXXXXXXKMFLSVEKENETIIKNQNHPK 704 + ++ LME++R Q ++ E++ + Sbjct: 270 ------------FFEVEALMEKMRQLQECREEEEASQEEMASRFEMEKRESMFVFSSDDD 317 Query: 705 EVNPL--IERFIEDPNFTYISYNSNNAEV--LRMQEYSWSDEACCLADRFYTDIGKALED 872 EV P + R ED ++ Y ++ + V R+Q+Y W D L +R Y D+G+ +++ Sbjct: 318 EVTPARDVSRHFEDTSYGYKDFSRHGMHVPTFRVQDYCWEDHGYSLVNRLYPDVGQLIDE 377 Query: 873 KFNCIIGKNI*YN 911 KF+ I N+ YN Sbjct: 378 KFH--IPYNLTYN 388
Score = 87.0 bits (214), Expect = 1e-16
Identities = 40/102 (39%), Positives = 66/102 (64%)
Frame = +1
Query: 898 TYNTMGDKTNIDTTIFRLSVWIYVHSLYGIRHDDFNYAQMDILLTNPLKTFLKKAMTKPC 1077
TYNTM ++DT++ R ++W Y+H ++GIR+DD++Y +++ LL K ++K + P
Sbjct: 386 TYNTMAMHKDVDTSMLRRAIWNYIHCMFGIRYDDYDYGEINQLLDRSFKVYIKTVVCTPE 445
Query: 1078 TSKKSDLFRFFPEFKISEKVHIALMAADARQQACLLHILKAM 1203
K F+ +FK SEKVH+ L+ +AR QA LL+ L+A+
Sbjct: 446 KVTKRMYDSFWRQFKHSEKVHVNLLLIEARMQAELLYALRAI 487
>sp|P58005|SESN3_HUMAN Sestrin-3 Length = 492 Score = 124 bits (312), Expect = 5e-28 Identities = 93/308 (30%), Positives = 144/308 (46%), Gaps = 5/308 (1%) Frame = +3 Query: 3 PYVDRFYLAIMAAAQMHCKELIYRFITEFIAAGGDPNWLQGLHKAPSRFRPLAVLNQHLC 182 PY R Y+AIMAAA+ C LI + EF+ GG WL GL P R + L +N+ L Sbjct: 106 PY--RHYIAIMAAARHQCSYLINMHVDEFLKTGGIAEWLNGLEYVPQRLKNLNEINKLLA 163 Query: 183 HQPWQIEPKNIQQLIHSPKGLFHISMSQLMQAVVIMTHVYSLSSFILVCGINSEIDHPPA 362 H+PW I ++IQ+L+ + G + S+ +L+ AVV++ H ++L+SF+ GIN E D P Sbjct: 164 HRPWLITKEHIQKLVKT--GENNWSLPELVHAVVLLAHYHALASFVFGSGINPERD-PEI 220 Query: 363 LLWYN--STNSDSSNSLRNTHSTENISLSNSNDVXXXXXXXXXXXXXXXXXXXXQSETDD 536 + S N+ L N ++ EN SLS SN Q E +D Sbjct: 221 SNGFRLISVNNFCVCDLANDNNIENASLSGSN--FGIVDSLSELEALMERMKRLQEERED 278 Query: 537 DKSDENHEDYPLDVLMERLRLAQXXXXXXXXXXXXKMFLSVEKENETIIKNQNHPKEVNP 716 +++ + + + R + F + E++ II + Sbjct: 279 EEASQ-------EEMSTRFEKEKKESLFVVSGDTFHSFPHSDFEDDMIITSD-------- 323 Query: 717 LIERFIEDPNFTYISYNSNNAEVL---RMQEYSWSDEACCLADRFYTDIGKALEDKFNCI 887 + R+IEDP F Y + E L R Q+Y+W + L +R Y+DIG L++KF + Sbjct: 324 -VSRYIEDPGFGYEDFARRGEEHLPTFRAQDYTWENHGFSLVNRLYSDIGHLLDEKFRMV 382 Query: 888 IGKNI*YN 911 N+ YN Sbjct: 383 Y--NLTYN 388
Score = 85.5 bits (210), Expect = 3e-16
Identities = 41/102 (40%), Positives = 67/102 (65%)
Frame = +1
Query: 898 TYNTMGDKTNIDTTIFRLSVWIYVHSLYGIRHDDFNYAQMDILLTNPLKTFLKKAMTKPC 1077
TYNTM ++DTT+ R +++ YVH ++GIR+DD++Y +++ LL LK ++K P
Sbjct: 386 TYNTMATHEDVDTTMLRRALFNYVHCMFGIRYDDYDYGEVNQLLERSLKVYIKTVTCYPE 445
Query: 1078 TSKKSDLFRFFPEFKISEKVHIALMAADARQQACLLHILKAM 1203
+ K ++ +FK SEKVH+ L+ +AR QA LL+ L+A+
Sbjct: 446 RTTKRMYDSYWRQFKHSEKVHVNLLLMEARMQAELLYALRAI 487
>sp|P58004|SESN2_HUMAN Sestrin-2 (Hi95) Length = 480 Score = 124 bits (311), Expect = 7e-28 Identities = 90/297 (30%), Positives = 138/297 (46%), Gaps = 9/297 (3%) Frame = +3 Query: 15 RFYLAIMAAAQMHCKELIYRFITEFIAAGGDPNWLQGLHKAPSRFRPLAVLNQHLCHQPW 194 R Y+AIMAAA+ C L+ + EF+ GGDP WL GLH+AP + R L+ +N+ L H+PW Sbjct: 112 RHYIAIMAAARHQCSYLVGSHMAEFLQTGGDPEWLLGLHRAPEKLRKLSEINKLLAHRPW 171 Query: 195 QIEPKNIQQLIHSPKGLFHISMSQLMQAVVIMTHVYSLSSFILVCGI--NSEIDHPPALL 368 I ++IQ L+ + G S+++L+QA+V++TH +SLSSF+ CGI + D PA Sbjct: 172 LITKEHIQALLKT--GEHTWSLAELIQALVLLTHCHSLSSFVFGCGILPEGDADGSPAPQ 229 Query: 369 WYNSTNSDSSNSLRNTHSTENISLSNSNDVXXXXXXXXXXXXXXXXXXXXQSETDDDKSD 548 + SS R+ L+NS +S D Sbjct: 230 APTPPSEQSSPPSRD-------PLNNSG--------------------GFESARD----- 257 Query: 549 ENHEDYPLDVLMERLRLAQXXXXXXXXXXXXKM--FLSVEKENETIIKNQNHPKEVNPLI 722 ++ LMER++ Q +M +EK ++ E +P Sbjct: 258 -------VEALMERMQQLQESLLRDEGTSQEEMESRFELEKSESLLVTPSADILEPSPHP 310 Query: 723 ER--FIEDPNFTYISYNSNNAE---VLRMQEYSWSDEACCLADRFYTDIGKALEDKF 878 + F+EDP F Y + A+ R Q+Y+W D L R Y + G+ L++KF Sbjct: 311 DMLCFVEDPTFGYEDFTRRGAQAPPTFRAQDYTWEDHGYSLIQRLYPEGGQLLDEKF 367
Score = 85.5 bits (210), Expect = 3e-16
Identities = 38/102 (37%), Positives = 66/102 (64%)
Frame = +1
Query: 898 TYNTMGDKTNIDTTIFRLSVWIYVHSLYGIRHDDFNYAQMDILLTNPLKTFLKKAMTKPC 1077
TYNT+ + +DT++ R ++W Y+H ++GIR+DD++Y +++ LL LK ++K P
Sbjct: 374 TYNTIAMHSGVDTSVLRRAIWNYIHCVFGIRYDDYDYGEVNQLLERNLKVYIKTVACYPE 433
Query: 1078 TSKKSDLFRFFPEFKISEKVHIALMAADARQQACLLHILKAM 1203
+ + F+ F+ SEKVH+ L+ +AR QA LL+ L+A+
Sbjct: 434 KTTRRMYNLFWRHFRHSEKVHVNLLLLEARMQAALLYALRAI 475
>sp|P58043|SESN2_MOUSE Sestrin-2 Length = 480 Score = 121 bits (304), Expect = 4e-27 Identities = 89/295 (30%), Positives = 141/295 (47%), Gaps = 7/295 (2%) Frame = +3 Query: 15 RFYLAIMAAAQMHCKELIYRFITEFIAAGGDPNWLQGLHKAPSRFRPLAVLNQHLCHQPW 194 R Y+AIMAAA+ C L+ +TEF+ GGDP WL GLH+AP + R L+ +N+ L H+PW Sbjct: 112 RHYIAIMAAARHQCSYLVGSHMTEFLQTGGDPEWLLGLHRAPEKLRKLSEVNKLLAHRPW 171 Query: 195 QIEPKNIQQLIHSPKGLFHISMSQLMQAVVIMTHVYSLSSFILVCGINSEIDHPPALLWY 374 I ++IQ L+ + G S+++L+QA+V++TH +SL+SF+ CGI E D + Sbjct: 172 LITKEHIQALLKT--GEHSWSLAELIQALVLLTHCHSLASFVFGCGILPEGDAEGS---- 225 Query: 375 NSTNSDSSNSLRNTHSTENISLSNSNDVXXXXXXXXXXXXXXXXXXXXQSETDDDKSDEN 554 ++ + S S + T + + L+NS ++ D Sbjct: 226 PASQAPSPPSEQGTPPSGD-PLNNSG--------------------GFEAARD------- 257 Query: 555 HEDYPLDVLMERLRLAQXXXXXXXXXXXXKM--FLSVEKENETIIKNQNHPKEVNPL--I 722 ++ LMER+R Q +M +EK ++ E +P I Sbjct: 258 -----VEALMERMRQLQESLLRDEGASQEEMENRFELEKSESLLVTPSADILEPSPHPDI 312 Query: 723 ERFIEDPNFTYISYNSNNAE---VLRMQEYSWSDEACCLADRFYTDIGKALEDKF 878 F+EDP F Y + + R Q+Y+W D L R Y + G+ L++KF Sbjct: 313 LCFVEDPAFGYEDFTRRGTQAPPTFRAQDYTWEDHGYSLIQRLYPEGGQLLDEKF 367
Score = 84.3 bits (207), Expect = 8e-16
Identities = 38/102 (37%), Positives = 66/102 (64%)
Frame = +1
Query: 898 TYNTMGDKTNIDTTIFRLSVWIYVHSLYGIRHDDFNYAQMDILLTNPLKTFLKKAMTKPC 1077
TYNT+ + +DT++ R ++W Y+H ++GIR+DD++Y +++ LL LK ++K P
Sbjct: 374 TYNTIAMHSGVDTSMLRRAIWNYIHCVFGIRYDDYDYGEVNQLLERNLKIYIKTVACYPE 433
Query: 1078 TSKKSDLFRFFPEFKISEKVHIALMAADARQQACLLHILKAM 1203
+ + F+ F+ SEKVH+ L+ +AR QA LL+ L+A+
Sbjct: 434 KTTRRMYNLFWRHFRHSEKVHVNLLLLEARMQAALLYALRAI 475
>sp|P58003|SESN1_XENLA Sestrin-1 (p53-regulated protein PA26) (XPA26) Length = 481 Score = 117 bits (293), Expect = 8e-26 Identities = 83/306 (27%), Positives = 136/306 (44%), Gaps = 9/306 (2%) Frame = +3 Query: 21 YLAIMAAAQMHCKELIYRFITEFIAAGGDPNWLQGLHKAPSRFRPLAVLNQHLCHQPWQI 200 Y+ IMAA++ C L+ + +FI GGD WL GL AP + R L LN+ L H+PW I Sbjct: 111 YIGIMAASRHQCSYLVNLHVNDFIHHGGDQKWLNGLENAPQKLRNLGELNKMLAHRPWLI 170 Query: 201 EPKNIQQLIHSPKGLFHISMSQLMQAVVIMTHVYSLSSFILVCGINSEID-------HPP 359 ++I+QL+ + G S+++L+ A+V++ H +SL+SF CGIN EI PP Sbjct: 171 TKEHIKQLLKA--GEHSWSLAELIHAIVLLAHYHSLASFTFGCGINPEIHSNGGHTFRPP 228 Query: 360 ALLWYNSTNSDSSNSLRNTHSTENISLSNSNDVXXXXXXXXXXXXXXXXXXXXQSETDDD 539 ++ Y + + N H I + + + + Sbjct: 229 SVSNYCICDITNGNHGTEGHLPVGIVMPTDS------------TCEVEALMVKMKQLQEG 276 Query: 540 KSDENHEDYPLDVLMERLRLAQXXXXXXXXXXXXKMFLSVEKENETIIKNQNHPKEVNPL 719 + +E + ER + M S E E+ P +++ Sbjct: 277 RDEEEASQEEMATRFEREK-------------TESMVFSTEDEDP--------PPDID-- 313 Query: 720 IERFIEDPNFTYISYNSNNAEV--LRMQEYSWSDEACCLADRFYTDIGKALEDKFNCIIG 893 + R ED ++ Y ++ V R+Q+YSW D L +R Y D+G+ L++KF+ I Sbjct: 314 VSRHFEDTSYGYKDFSRRGMHVPTFRVQDYSWEDHGYSLVNRLYPDVGQLLDEKFH--IA 371 Query: 894 KNI*YN 911 N+ YN Sbjct: 372 YNLTYN 377
Score = 87.4 bits (215), Expect = 9e-17
Identities = 41/102 (40%), Positives = 65/102 (63%)
Frame = +1
Query: 898 TYNTMGDKTNIDTTIFRLSVWIYVHSLYGIRHDDFNYAQMDILLTNPLKTFLKKAMTKPC 1077
TYNTM ++DT+ R +VW YVH ++GIR+DD++Y +++ LL K ++K + P
Sbjct: 375 TYNTMAMHKDVDTSTLRRAVWNYVHCMFGIRYDDYDYGEINQLLDRSFKVYIKNVVCSPE 434
Query: 1078 TSKKSDLFRFFPEFKISEKVHIALMAADARQQACLLHILKAM 1203
+ K F+ +FK SEKVH+ L+ +AR Q LL+ L+A+
Sbjct: 435 KTSKRMYDGFWRQFKHSEKVHVNLLLMEARMQGELLYALRAI 476
>sp|Q9W1K5|SEST_DROME Putative sestrin Length = 497 Score = 115 bits (287), Expect = 4e-25 Identities = 83/315 (26%), Positives = 139/315 (44%), Gaps = 27/315 (8%) Frame = +3 Query: 15 RFYLAIMAAAQMHCKELIYRFITEFIAAGGDPNWLQGLHKAPSRFRPLAVLNQHLCHQPW 194 R+YLAI+AAA+ C L+ R+ EFI GGD WL GL P++ R + +N+ L H+PW Sbjct: 73 RYYLAIIAAARHQCPYLVKRYEKEFINQGGDSAWLGGLDFIPAKLRAIYDINKILAHRPW 132 Query: 195 QIEPKNIQQLIHSPKGLFHISMSQLMQAVVIMTHVYSLSSFILVCGINSEID-------- 350 + ++I++L KG S+S+++ A+V+++H +SLSSF+ CG+ ++D Sbjct: 133 LLRKEHIERL---TKGKNSWSLSEVVHAMVLLSHFHSLSSFVFSCGLTQKLDGLSSPKLK 189 Query: 351 HPPALLWYNS-----TNSDSSNSLRNTHSTENISLSNSNDVXXXXXXXXXXXXXXXXXXX 515 PPA + + T + + + ISL+N+N Sbjct: 190 SPPAAVAALAPTILITPTSPTEPQKGKPVLAEISLNNANPDYDSQTAASSNGGAPPDSAN 249 Query: 516 XQSETDDDKSDENH----------EDYPLDVLMERLRLAQXXXXXXXXXXXXKMFLSVEK 665 ++ D + + ++ LMER+++ F VE+ Sbjct: 250 AVADGPDATTLNGYLATAQQLPQQHGISVETLMERMKVLSQKQDECSEAELSSRFQKVEQ 309 Query: 666 EN-ETIIKNQNHPKEVNPLIERFIEDPNFTYISYNSNNAE---VLRMQEYSWSDEACCLA 833 + E V + +++D NF Y + E R+Q+YSW D L Sbjct: 310 QTAELAAVTPEAAVGVPTNLSHYVDDANFIYQDFARRGTESINTFRIQDYSWEDHGYSLV 369 Query: 834 DRFYTDIGKALEDKF 878 D Y D+G L+ KF Sbjct: 370 DGLYNDVGIFLDAKF 384
Score = 90.1 bits (222), Expect = 1e-17
Identities = 45/100 (45%), Positives = 62/100 (62%)
Frame = +1
Query: 898 TYNTMGDKTNIDTTIFRLSVWIYVHSLYGIRHDDFNYAQMDILLTNPLKTFLKKAMTKPC 1077
TY TMG N+DT+ FR ++W Y+ +YGIRHDD++Y +++ LL PLK F+K A P
Sbjct: 391 TYCTMGGIKNVDTSKFRRAIWNYIQCIYGIRHDDYDYGEVNQLLVRPLKMFIKTACCFPE 450
Query: 1078 TSKKSDLFRFFPEFKISEKVHIALMAADARQQACLLHILK 1197
D E + SEKVH+ LM +AR QA LL+ L+
Sbjct: 451 RITTKDYDSVLVELQDSEKVHVNLMIMEARNQAELLYALR 490
>sp|Q9N4D6|SEST_CAEEL Putative sestrin Length = 517 Score = 74.3 bits (181), Expect = 8e-13 Identities = 37/92 (40%), Positives = 56/92 (60%) Frame = +1 Query: 928 IDTTIFRLSVWIYVHSLYGIRHDDFNYAQMDILLTNPLKTFLKKAMTKPCTSKKSDLFRF 1107 IDT FR ++W Y LYG+R DD++Y++++ +L KTF+K A P ++ R Sbjct: 409 IDTAAFREAIWNYTQGLYGVRVDDYDYSKINRVLDKGTKTFIKLAACYP-HKLTTEFTRA 467 Query: 1108 FPEFKISEKVHIALMAADARQQACLLHILKAM 1203 P FK SEK+H+ +M A AR QA + H +A+ Sbjct: 468 LPGFKDSEKIHVVMMVAMARFQASMFHYTRAV 499
Score = 68.9 bits (167), Expect = 3e-11
Identities = 41/119 (34%), Positives = 62/119 (52%), Gaps = 3/119 (2%)
Frame = +3
Query: 3 PYVDRFYLAIMAAAQMHCKELIYRFITEFIAAGGDPNWLQGLHKAPSRFRPLAVLNQHLC 182
P R YLA+MAA++ C L+ +F+ GG WL G+ + +FR L LN+ LC
Sbjct: 115 PASARHYLALMAASRHRCLSLMDHHRRKFLEKGGQQMWLLGIDWSYMKFRKLDYLNRMLC 174
Query: 183 HQPWQIEPKNIQQLI--HSPK-GLFHISMSQLMQAVVIMTHVYSLSSFILVCGINSEID 350
H+PW I KN+ L +P G S+ L AV IM+ +++ + I G+ +D
Sbjct: 175 HRPWMIHWKNLAVLFARRTPDGGPAFFSVCSLHHAVGIMSMTHAMCTVICCIGLERRVD 233
>sp|Q9Y7J8|YGL1_SCHPO Hypothetical protein C216.01c in chromosome II Length = 836 Score = 38.1 bits (87), Expect = 0.064 Identities = 33/94 (35%), Positives = 46/94 (48%), Gaps = 16/94 (17%) Frame = +3 Query: 213 IQQLIHSPKGLFHISMSQLMQAVV----------IMTH----VYSLSSFILVCGINSEID 350 IQQ+ + KGL S S L V M+H V + S ILV S ID Sbjct: 373 IQQVCNIAKGLQFQSCSALFATFVKFNLLKALDYAMSHENNSVRNAGSDILV----SIID 428 Query: 351 HPPALLW--YNSTNSDSSNSLRNTHSTENISLSN 446 H PA++W ++ D+S+SL N H +++ LSN Sbjct: 429 HEPAIVWQKFDQDRKDASSSLSNAHVSQHSLLSN 462
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 147,194,270
Number of Sequences: 369166
Number of extensions: 2931609
Number of successful extensions: 7133
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6699
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7115
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 17168153970
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail