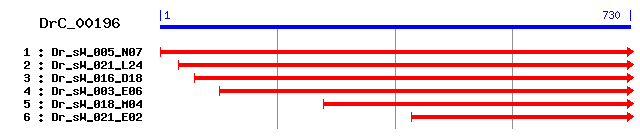
DrC_00196
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00196 (730 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q24400|MLP2_DROME Muscle LIM protein Mlp84B 157 3e-38 sp|P67966|CSRP1_CHICK Cysteine-rich protein 1 (CRP1) (CRP) ... 124 2e-28 sp|P47875|CSRP1_RAT Cysteine and glycine-rich protein 1 (Cy... 120 5e-27 sp|Q62908|CSRP2_RAT Cysteine and glycine-rich protein 2 (Cy... 119 8e-27 sp|Q05158|CSRP2_COTJA Cysteine and glycine-rich protein 2 (... 119 8e-27 sp|P50460|CSRP2_CHICK Cysteine and glycine-rich protein 2 (... 119 1e-26 sp|Q3MHY1|CSRP1_BOVIN Cysteine and glycine-rich protein 1 (... 119 1e-26 sp|P97314|CSRP2_MOUSE Cysteine and glycine-rich protein 2 (... 118 1e-26 sp|Q16527|CSRP2_HUMAN Cysteine and glycine-rich protein 2 (... 118 1e-26 sp|P97315|CSRP1_MOUSE Cysteine and glycine-rich protein 1 (... 117 2e-26
>sp|Q24400|MLP2_DROME Muscle LIM protein Mlp84B Length = 495 Score = 157 bits (397), Expect = 3e-38 Identities = 81/168 (48%), Positives = 99/168 (58%), Gaps = 7/168 (4%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 CP+C K+VYAAE++LAG V+H C KCGMCNK LDST EH+ ELYCK CH Sbjct: 12 CPRCGKSVYAAEERLAGGYVFHKNCFKCGMCNKSLDSTNCTEHERELYCKTCHGRKFGPK 71 Query: 266 XXXXXXXXXTLNMDKGGQFGNKDAPTGNKPNVMGGKVV-------APEDGGCPRCGKRVY 424 TL+MD G QF ++ G+ P+V G + APE GCPRCG VY Sbjct: 72 GYGFGTGAGTLSMDNGSQFLREN---GDVPSVRNGARLEPRAIARAPEGEGCPRCGGYVY 128 Query: 425 DAEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 AE+ + G S+HK CF C CKK LDS ILC +K++YCK CY Sbjct: 129 AAEQMLA--RGRSWHKECFKCGTCKKGLDS-ILCCEAPDKNIYCKGCY 173
Score = 114 bits (284), Expect = 3e-25
Identities = 63/164 (38%), Positives = 83/164 (50%), Gaps = 1/164 (0%)
Frame = +2
Query: 80 EICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAE-HDNELYCKQCHXXXX 256
E CP+C VYAAE+ LA + WH C KCG C K LDS E D +YCK C+
Sbjct: 118 EGCPRCGGYVYAAEQMLARGRSWHKECFKCGTCKKGLDSILCCEAPDKNIYCKGCYAKKF 177
Query: 257 XXXXXXXXXXXXTLNMDKGGQFGNKDAPTGNKPNVMGGKVVAPEDGGCPRCGKRVYDAEK 436
L D + + D + + K+ A GCPRCG VY AE+
Sbjct: 178 GPKGYGYGQGGGALQSD---CYAHDDGAPQIRAAIDVDKIQARPGEGCPRCGGVVYAAEQ 234
Query: 437 AIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568
+ G +HK CF+CK+C K LDS I ++ ++DVYC+TCY
Sbjct: 235 KLS--KGREWHKKCFNCKDCHKTLDS-INASDGPDRDVYCRTCY 275
Score = 101 bits (252), Expect = 2e-21
Identities = 54/166 (32%), Positives = 84/166 (50%), Gaps = 3/166 (1%)
Frame = +2
Query: 80 EICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEH-DNELYCKQCHXXXX 256
E CP+C VYAAE+KL+ + WH C C C+K LDS ++ D ++YC+ C+
Sbjct: 220 EGCPRCGGVVYAAEQKLSKGREWHKKCFNCKDCHKTLDSINASDGPDRDVYCRTCYGKKW 279
Query: 257 XXXXXXXXXXXXTLNMDKGGQFGNKDAPTGNKP--NVMGGKVVAPEDGGCPRCGKRVYDA 430
L D +D + N+P N + A + GCPRCG V+ A
Sbjct: 280 GPHGYGFACGSGFLQTDGL----TEDQISANRPFYNPDTTSIKARDGEGCPRCGGAVFAA 335
Query: 431 EKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568
E+ + G +HK C++C +C + LDS + C + + D++C+ CY
Sbjct: 336 EQQLS--KGKVWHKKCYNCADCHRPLDSVLAC-DGPDGDIHCRACY 378
Score = 101 bits (251), Expect = 2e-21
Identities = 59/177 (33%), Positives = 86/177 (48%), Gaps = 14/177 (7%)
Frame = +2
Query: 80 EICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAE-HDNELYCKQCHXXXX 256
E CP+C AV+AAE++L+ KVWH C C C++ LDS + D +++C+ C+
Sbjct: 323 EGCPRCGGAVFAAEQQLSKGKVWHKKCYNCADCHRPLDSVLACDGPDGDIHCRACY---- 378
Query: 257 XXXXXXXXXXXXTLNMDKGGQFGNKDAPTGNKPNVM------------GGKVVAPE-DGG 397
G FG K G+ P ++ G + P+ GG
Sbjct: 379 ------------------GKLFGPKGFGYGHAPTLVSTSGESTIQFPDGRPLAGPKTSGG 420
Query: 398 CPRCGKRVYDAEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568
CPRCG V+ AE+ I +HK CF C +C+K LDST L + + D+YC+ CY
Sbjct: 421 CPRCGFAVFAAEQMIS--KTRIWHKRCFYCSDCRKSLDSTNL-NDGPDGDIYCRACY 474
Score = 62.0 bits (149), Expect = 2e-09
Identities = 25/60 (41%), Positives = 38/60 (63%), Gaps = 1/60 (1%)
Frame = +2
Query: 68 PKMSEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAE-HDNELYCKQCH 244
PK S CP+C AV+AAE+ ++ ++WH C C C K LDST + + D ++YC+ C+
Sbjct: 415 PKTSGGCPRCGFAVFAAEQMISKTRIWHKRCFYCSDCRKSLDSTNLNDGPDGDIYCRACY 474
>sp|P67966|CSRP1_CHICK Cysteine-rich protein 1 (CRP1) (CRP) sp|P67967|CSRP1_COTJA Cysteine-rich protein 1 (CRP1) (CRP) Length = 192 Score = 124 bits (312), Expect = 2e-28 Identities = 69/167 (41%), Positives = 88/167 (52%), Gaps = 6/167 (3%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 C C+KAVY AE+ +H C C +C K LDSTTVA H +E+YCK C+ Sbjct: 10 CGVCQKAVYFAEEVQCEGSSFHKSCFLCMVCKKNLDSTTVAVHGDEIYCKSCYGKKYGPK 69 Query: 266 XXXXXXXXXTLNMDKGGQFGNK------DAPTGNKPNVMGGKVVAPEDGGCPRCGKRVYD 427 TL+ DKG G K PT + M KV + GCPRCG+ VY Sbjct: 70 GYGYGMGAGTLSTDKGESLGIKYEEGQSHRPTNPNASRMAQKVGGSD--GCPRCGQAVYA 127 Query: 428 AEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 AEK IG G S+HK+CF C +C K L+ST L ++ ++YCK CY Sbjct: 128 AEKVIGA--GKSWHKSCFRCAKCGKSLESTTLA--DKDGEIYCKGCY 170
Score = 75.1 bits (183), Expect = 2e-13
Identities = 29/56 (51%), Positives = 39/56 (69%)
Frame = +2
Query: 77 SEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCH 244
S+ CP+C +AVYAAEK + K WH C +C C K L+STT+A+ D E+YCK C+
Sbjct: 115 SDGCPRCGQAVYAAEKVIGAGKSWHKSCFRCAKCGKSLESTTLADKDGEIYCKGCY 170
>sp|P47875|CSRP1_RAT Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP) Length = 193 Score = 120 bits (300), Expect = 5e-27 Identities = 68/167 (40%), Positives = 90/167 (53%), Gaps = 6/167 (3%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 C C+K VY AE+ +H C C +C K LDSTTVA H E+YCK C+ Sbjct: 10 CGVCQKTVYFAEEVQCEGNSFHKSCFLCMVCKKNLDSTTVAVHGEEIYCKSCYGKKYGPK 69 Query: 266 XXXXXXXXXTLNMDKGGQFG--NKDAPTGNKPNV-MGGKVVAPEDGG---CPRCGKRVYD 427 TL+MDKG G +++AP G++P A + GG CPRC + VY Sbjct: 70 GYGYGQGAGTLSMDKGESLGIKHEEAP-GHRPTTNPNASKFAQKIGGSERCPRCSQAVYA 128 Query: 428 AEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 AEK IG G S+HK+CF C +C K L+ST L ++ ++YCK CY Sbjct: 129 AEKVIGA--GKSWHKSCFRCAKCGKGLESTTLA--DKDGEIYCKGCY 171
Score = 76.6 bits (187), Expect = 6e-14
Identities = 30/56 (53%), Positives = 39/56 (69%)
Frame = +2
Query: 77 SEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCH 244
SE CP+C +AVYAAEK + K WH C +C C K L+STT+A+ D E+YCK C+
Sbjct: 116 SERCPRCSQAVYAAEKVIGAGKSWHKSCFRCAKCGKGLESTTLADKDGEIYCKGCY 171
>sp|Q62908|CSRP2_RAT Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Smooth muscle cell LIM protein) (SmLIM) Length = 193 Score = 119 bits (298), Expect = 8e-27 Identities = 70/170 (41%), Positives = 87/170 (51%), Gaps = 9/170 (5%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 C C + VY AE+ + +H C C +C K LDSTTVA HD E+YCK C+ Sbjct: 10 CGACGRTVYHAEEVQCDGRTFHRCCFLCMVCRKNLDSTTVAIHDEEIYCKSCYGKKYGPK 69 Query: 266 XXXXXXXXXTLNMDKGGQFGNK------DAPTGNKPNVMGGKVVAPEDGG---CPRCGKR 418 TLNMD+G + G K PT N PN A + GG C RCG Sbjct: 70 GYGYGQGAGTLNMDRGERLGIKPESAQPHRPTTN-PNT---SKFAQKYGGAEKCSRCGDS 125 Query: 419 VYDAEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 VY AEK IG G +HK CF C +C K L+ST L ++E ++YCK CY Sbjct: 126 VYAAEKIIGA--GKPWHKNCFRCAKCGKSLESTTL--TEKEGEIYCKGCY 171
Score = 67.8 bits (164), Expect = 3e-11
Identities = 26/56 (46%), Positives = 36/56 (64%)
Frame = +2
Query: 77 SEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCH 244
+E C +C +VYAAEK + K WH C +C C K L+STT+ E + E+YCK C+
Sbjct: 116 AEKCSRCGDSVYAAEKIIGAGKPWHKNCFRCAKCGKSLESTTLTEKEGEIYCKGCY 171
>sp|Q05158|CSRP2_COTJA Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) Length = 194 Score = 119 bits (298), Expect = 8e-27 Identities = 67/167 (40%), Positives = 88/167 (52%), Gaps = 6/167 (3%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 C C + VY AE+ + +H C C +C K LDSTTVA HD E+YCK C+ Sbjct: 10 CGACGRTVYHAEEVQCDGRSFHRCCFLCMVCRKNLDSTTVAIHDAEVYCKSCYGKKYGPK 69 Query: 266 XXXXXXXXXTLNMDKGGQFGNK--DAPTGNKPNV-MGGKVVAPEDGG---CPRCGKRVYD 427 TLNMD+G + G K +P+ ++P A + GG C RCG VY Sbjct: 70 GYGYGQGAGTLNMDRGERLGIKPESSPSPHRPTTNPNTSKFAQKFGGAEKCSRCGDSVYA 129 Query: 428 AEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 AEK IG G +HK CF C +C K L+ST L ++E ++YCK CY Sbjct: 130 AEKVIGA--GKPWHKNCFRCAKCGKSLESTTL--TEKEGEIYCKGCY 172
Score = 68.2 bits (165), Expect = 2e-11
Identities = 26/56 (46%), Positives = 36/56 (64%)
Frame = +2
Query: 77 SEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCH 244
+E C +C +VYAAEK + K WH C +C C K L+STT+ E + E+YCK C+
Sbjct: 117 AEKCSRCGDSVYAAEKVIGAGKPWHKNCFRCAKCGKSLESTTLTEKEGEIYCKGCY 172
>sp|P50460|CSRP2_CHICK Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Beta-cysteine-rich protein) (Beta-CRP) Length = 194 Score = 119 bits (297), Expect = 1e-26 Identities = 67/167 (40%), Positives = 87/167 (52%), Gaps = 6/167 (3%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 C C + VY AE+ + +H C C +C K LDSTTVA HD E+YCK C+ Sbjct: 10 CGACGRTVYHAEEVQCDGRSFHRCCFLCMVCRKNLDSTTVAIHDAEVYCKSCYGKKYGPK 69 Query: 266 XXXXXXXXXTLNMDKGGQFGNK--DAPTGNKPNV-MGGKVVAPEDGG---CPRCGKRVYD 427 TLNMD+G + G K P+ ++P A + GG C RCG VY Sbjct: 70 GYGYGQGAGTLNMDRGERLGIKPESTPSPHRPTTNPNTSKFAQKFGGAEKCSRCGDSVYA 129 Query: 428 AEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 AEK IG G +HK CF C +C K L+ST L ++E ++YCK CY Sbjct: 130 AEKVIGA--GKPWHKNCFRCAKCGKSLESTTL--TEKEGEIYCKGCY 172
Score = 68.2 bits (165), Expect = 2e-11
Identities = 26/56 (46%), Positives = 36/56 (64%)
Frame = +2
Query: 77 SEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCH 244
+E C +C +VYAAEK + K WH C +C C K L+STT+ E + E+YCK C+
Sbjct: 117 AEKCSRCGDSVYAAEKVIGAGKPWHKNCFRCAKCGKSLESTTLTEKEGEIYCKGCY 172
>sp|Q3MHY1|CSRP1_BOVIN Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) Length = 193 Score = 119 bits (297), Expect = 1e-26 Identities = 68/167 (40%), Positives = 90/167 (53%), Gaps = 6/167 (3%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 C C+K VY AE+ +H C C +C K LDSTTVA H E+YCK C+ Sbjct: 10 CGVCQKTVYFAEEVQCEGSSFHKSCFLCLVCKKNLDSTTVAVHGEEIYCKSCYGKKYGPK 69 Query: 266 XXXXXXXXXTLNMDKGGQFG--NKDAPTGNKPNV-MGGKVVAPEDGG---CPRCGKRVYD 427 TL+MDKG G +++AP G++P A + GG CPRC + VY Sbjct: 70 GYGYGQGAGTLSMDKGESLGIRHEEAP-GHRPTTNPNTSKFAQKVGGSERCPRCSQAVYA 128 Query: 428 AEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 AEK IG G S+HK+CF C +C K L+ST L ++ ++YCK CY Sbjct: 129 AEKVIGA--GKSWHKSCFRCAKCGKGLESTTLA--DKDGEIYCKGCY 171
Score = 76.6 bits (187), Expect = 6e-14
Identities = 30/56 (53%), Positives = 39/56 (69%)
Frame = +2
Query: 77 SEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCH 244
SE CP+C +AVYAAEK + K WH C +C C K L+STT+A+ D E+YCK C+
Sbjct: 116 SERCPRCSQAVYAAEKVIGAGKSWHKSCFRCAKCGKGLESTTLADKDGEIYCKGCY 171
>sp|P97314|CSRP2_MOUSE Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Double LIM protein 1) (DLP-1) Length = 193 Score = 118 bits (296), Expect = 1e-26 Identities = 70/170 (41%), Positives = 87/170 (51%), Gaps = 9/170 (5%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 C C + VY AE+ + +H C C +C K LDSTTVA HD E+YCK C+ Sbjct: 10 CGACGRTVYHAEEVQCDGRSFHRCCFLCMVCRKNLDSTTVAIHDEEIYCKSCYGKKYGPK 69 Query: 266 XXXXXXXXXTLNMDKGGQFGNK------DAPTGNKPNVMGGKVVAPEDGG---CPRCGKR 418 TLNMD+G + G K PT N PN A + GG C RCG Sbjct: 70 GYGYGQGAGTLNMDRGERLGIKPESAQPHRPTTN-PNT---SKFAQKYGGAEKCSRCGDS 125 Query: 419 VYDAEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 VY AEK IG G +HK CF C +C K L+ST L ++E ++YCK CY Sbjct: 126 VYAAEKIIGA--GKPWHKNCFRCAKCGKSLESTTL--TEKEGEIYCKGCY 171
Score = 67.8 bits (164), Expect = 3e-11
Identities = 26/56 (46%), Positives = 36/56 (64%)
Frame = +2
Query: 77 SEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCH 244
+E C +C +VYAAEK + K WH C +C C K L+STT+ E + E+YCK C+
Sbjct: 116 AEKCSRCGDSVYAAEKIIGAGKPWHKNCFRCAKCGKSLESTTLTEKEGEIYCKGCY 171
>sp|Q16527|CSRP2_HUMAN Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Smooth muscle cell LIM protein) (SmLIM) (LIM-only protein 5) Length = 193 Score = 118 bits (296), Expect = 1e-26 Identities = 70/170 (41%), Positives = 87/170 (51%), Gaps = 9/170 (5%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 C C + VY AE+ + +H C C +C K LDSTTVA HD E+YCK C+ Sbjct: 10 CGACGRTVYHAEEVQCDGRSFHRCCFLCMVCRKNLDSTTVAIHDEEIYCKSCYGKKYGPK 69 Query: 266 XXXXXXXXXTLNMDKGGQFGNK------DAPTGNKPNVMGGKVVAPEDGG---CPRCGKR 418 TLNMD+G + G K PT N PN A + GG C RCG Sbjct: 70 GYGYGQGAGTLNMDRGERLGIKPESVQPHRPTTN-PNT---SKFAQKYGGAEKCSRCGDS 125 Query: 419 VYDAEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 VY AEK IG G +HK CF C +C K L+ST L ++E ++YCK CY Sbjct: 126 VYAAEKIIGA--GKPWHKNCFRCAKCGKSLESTTL--TEKEGEIYCKGCY 171
Score = 67.8 bits (164), Expect = 3e-11
Identities = 26/56 (46%), Positives = 36/56 (64%)
Frame = +2
Query: 77 SEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCH 244
+E C +C +VYAAEK + K WH C +C C K L+STT+ E + E+YCK C+
Sbjct: 116 AEKCSRCGDSVYAAEKIIGAGKPWHKNCFRCAKCGKSLESTTLTEKEGEIYCKGCY 171
>sp|P97315|CSRP1_MOUSE Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP) Length = 193 Score = 117 bits (294), Expect = 2e-26 Identities = 67/167 (40%), Positives = 89/167 (53%), Gaps = 6/167 (3%) Frame = +2 Query: 86 CPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCHXXXXXXX 265 C C+K VY AE+ +H C C +C K LDSTTVA H E+YCK C+ Sbjct: 10 CGVCQKTVYFAEEVQCEGNSFHKSCFLCMVCKKNLDSTTVAVHGEEIYCKSCYGKKYGPK 69 Query: 266 XXXXXXXXXTLNMDKGGQFG--NKDAPTGNKPNV-MGGKVVAPEDGG---CPRCGKRVYD 427 TL+ DKG G +++AP G++P A + GG CPRC + VY Sbjct: 70 GYGYGQGAGTLSTDKGESLGIKHEEAP-GHRPTTNPNASKFAQKIGGSERCPRCSQAVYA 128 Query: 428 AEKAIGCPDGLSFHKTCFSCKECKKKLDSTILCTNKEEKDVYCKTCY 568 AEK IG G S+HK+CF C +C K L+ST L ++ ++YCK CY Sbjct: 129 AEKVIGA--GKSWHKSCFRCAKCGKGLESTTLA--DKDGEIYCKGCY 171
Score = 76.6 bits (187), Expect = 6e-14
Identities = 30/56 (53%), Positives = 39/56 (69%)
Frame = +2
Query: 77 SEICPKCKKAVYAAEKKLAGNKVWHSMCLKCGMCNKMLDSTTVAEHDNELYCKQCH 244
SE CP+C +AVYAAEK + K WH C +C C K L+STT+A+ D E+YCK C+
Sbjct: 116 SERCPRCSQAVYAAEKVIGAGKSWHKSCFRCAKCGKGLESTTLADKDGEIYCKGCY 171
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 83,422,150
Number of Sequences: 369166
Number of extensions: 1741645
Number of successful extensions: 6368
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5581
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6221
length of database: 68,354,980
effective HSP length: 108
effective length of database: 48,403,600
effective search space used: 6486082400
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail