DrC_00129
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00129 (933 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P36104|SWD2_YEAST COMPASS component SWD2 (Complex protei... 171 3e-42 sp|O60137|SWD2_SCHPO Set1 complex component swd2 (Set1C com... 158 2e-38 sp|Q8YTC2|Y2800_ANASP Hypothetical WD-repeat protein alr2800 90 1e-17 sp|Q8YRI1|YY46_ANASP Hypothetical WD-repeat protein alr3466 88 4e-17 sp|P49695|PKWA_THECU Probable serine/threonine-protein kina... 86 1e-16 sp|Q9V3J8|WDS_DROME Will die slowly protein 86 1e-16 sp|Q17963|YKY4_CAEEL Hypothetical WD-repeat protein C14B1.4... 84 4e-16 sp|P61964|WDR5_HUMAN WD-repeat protein 5 (BMP2-induced 3-kb... 84 7e-16 sp|Q00808|HETE1_PODAN Vegetatible incompatibility protein H... 78 3e-14 sp|Q8YV57|Y2124_ANASP Hypothetical WD-repeat protein all2124 77 9e-14
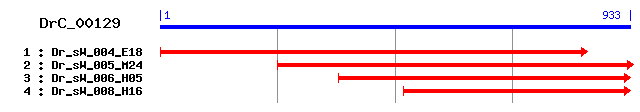
>sp|P36104|SWD2_YEAST COMPASS component SWD2 (Complex proteins associated with SET1 protein SWD2) (Set1C component SWD2) Length = 329 Score = 171 bits (433), Expect = 3e-42 Identities = 102/286 (35%), Positives = 158/286 (55%), Gaps = 7/286 (2%) Frame = +1 Query: 4 LISSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLH 183 L++SS +D M +Y + SKKYG + FTH+ + I++ST + I++L+L Sbjct: 42 LLTSSSNDTMQLYSATNCKFLDTIASKKYGCHSAIFTHAQNECIYSSTMKNFDIKYLNLE 101 Query: 184 DNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVGAF 363 N+Y+RYF GH V L M+PVND FLS S D+++RLWDLK Q I + A+ Sbjct: 102 TNQYLRYFSGHGALVNDLKMNPVNDTFLSSSYDESVRLWDLKISKPQVIIPSLVPNCIAY 161 Query: 364 DPEGQIFVAGVNSESMR--LYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRMIAICS 537 DP G +F G N E+ LY+++ +GPF+ + +WN L+FS NG+ + + S Sbjct: 162 DPSGLVFALG-NPENFEIGLYNLKKIQEGPFLIIKINDATFSQWNKLEFSNNGKYLLVGS 220 Query: 538 NGSQIKIIDAFDGKQLSNFMILGANPQRVPID---VCFSPDSQYLVCGAPDGQLCIWNTE 708 + + I DAF G+QL + A P R +D CF+PD ++++ DG++ IWN Sbjct: 221 SIGKHLIFDAFTGQQLFELIGTRAFPMREFLDSGSACFTPDGEFVLGTDYDGRIAIWNHS 280 Query: 709 N--KNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSAVTNLTFW 840 + N+V GF + + P +AFNPKY+M +A + F+ Sbjct: 281 DSISNKVLRPQGFIPCVSHETCP-RSIAFNPKYSMFVTADETVDFY 325
>sp|O60137|SWD2_SCHPO Set1 complex component swd2 (Set1C component swd2) (COMPASS component swd2) (Complex proteins associated with set1 protein swd2) Length = 357 Score = 158 bits (400), Expect = 2e-38 Identities = 99/322 (30%), Positives = 165/322 (51%), Gaps = 43/322 (13%) Frame = +1 Query: 7 ISSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLHD 186 ++S DD + +Y+C++G + L SKKYG +L +FTH S++ IHASTK D+T+R+L + Sbjct: 42 LASCTDDTLQLYNCISGKFVKSLASKKYGAHLGRFTHHSNSLIHASTKEDNTVRYLDVVT 101 Query: 187 NKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVGAFD 366 N+Y+RYF GH + V ++ +SP ++ FLS SLD TIRLWDL++ N QG ++ + V AFD Sbjct: 102 NRYLRYFPGHKQTVTSIDVSPADETFLSASLDNTIRLWDLRSPNCQGLLNVSSPVVAAFD 161 Query: 367 PEGQIFVA-GVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNA-LKFSPNGRMIAICSN 540 G IF + + LY+I+++D PF + A ++FS +G+ + + + Sbjct: 162 ATGLIFASVSERKYKISLYNIKSFDARPFQDIPLTFLPPHVRIANVEFSTDGKYLLLTTG 221 Query: 541 GSQIKIIDAFDGKQLSNFMILGANPQRVP-----------IDVCFSPDSQYLVCGAPDGQ 687 +IDA+ G +L P +V C SPDS++L + Sbjct: 222 NDFHYVIDAYSGSELLRV------PSKVSTKTQDGNLTYYASACMSPDSKFLFTSYDNEH 275 Query: 688 LCIWN----TENKNRVALVAGFDH---------------------TPNSTSQPI-----N 777 LC++ + N +V F P+STS+ + + Sbjct: 276 LCMYQLPELKQTVNSDHIVNSFTEMGLASTTPTSNSHSTENPLAIIPSSTSKNLLPETPS 335 Query: 778 CVAFNPKYAMISSAVTNLTFWS 843 V FNP+++ + +A + + FW+ Sbjct: 336 IVRFNPRFSQLVTAHSGVIFWT 357
>sp|Q8YTC2|Y2800_ANASP Hypothetical WD-repeat protein alr2800 Length = 1258 Score = 89.7 bits (221), Expect = 1e-17 Identities = 76/280 (27%), Positives = 131/280 (46%), Gaps = 3/280 (1%) Frame = +1 Query: 4 LISSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLH 183 L SS+ D + ++D G R L S V + F+ T AS D TI+ + H Sbjct: 783 LASSAADHTIKLWDVSQGKCLRTLKSHTGWVRSVAFSADGQTL--ASGSGDRTIKIWNYH 840 Query: 184 DNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPV--G 357 + ++ ++GHT V +++ SP + + +SGS D+TI+LWD + H +H V Sbjct: 841 TGECLKTYIGHTNSVYSIAYSPDSKILVSGSGDRTIKLWDCQTHICIKTLHGHTNEVCSV 900 Query: 358 AFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWN-ALKFSPNGRMIAIC 534 AF P+GQ +S+RL++ R G + +W + FSP+ +++A Sbjct: 901 AFSPDGQTLACVSLDQSVRLWNCRT---GQCLK---AWYGNTDWALPVAFSPDRQILASG 954 Query: 535 SNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWNTENK 714 SN +K+ D GK +S+ L + + + FSPDSQ L + D + +WN Sbjct: 955 SNDKTVKLWDWQTGKYISS---LEGHTDFI-YGIAFSPDSQTLASASTDSSVRLWNISTG 1010 Query: 715 NRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSAVTNLT 834 ++ +HT + V F+P+ +I++ + T Sbjct: 1011 QCFQIL--LEHT-----DWVYAVVFHPQGKIIATGSADCT 1043
Score = 78.2 bits (191), Expect = 3e-14
Identities = 71/289 (24%), Positives = 122/289 (42%), Gaps = 4/289 (1%)
Frame = +1
Query: 4 LISSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLH 183
L S S D + ++D TG L + + F+ S T AST D ++R ++
Sbjct: 951 LASGSNDKTVKLWDWQTGKYISSLEGHTDFIYGIAFSPDSQTLASAST--DSSVRLWNIS 1008
Query: 184 DNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFI--HTTGRPVG 357
+ + + HT V + P + +GS D T++LW++ + H+
Sbjct: 1009 TGQCFQILLEHTDWVYAVVFHPQGKIIATGSADCTVKLWNISTGQCLKTLSEHSDKILGM 1068
Query: 358 AFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRMIAICS 537
A+ P+GQ+ + +S+RL+D G V +++ + + FSPNG +IA CS
Sbjct: 1069 AWSPDGQLLASASADQSVRLWDCCT---GRCV--GILRGHSNRVYSAIFSPNGEIIATCS 1123
Query: 538 NGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWNTENKN 717
+KI D GK L D+ FSPD + L + D + IW+
Sbjct: 1124 TDQTVKIWDWQQGKCLKTL----TGHTNWVFDIAFSPDGKILASASHDQTVRIWDVNTGK 1179
Query: 718 RVALVAGFDHTPNSTSQPINCVAFNPKYAMISSAVTNLT--FWSPQIDE 858
+ G H ++ VAF+P +++S + T W+ + E
Sbjct: 1180 CHHICIGHTHL-------VSSVAFSPDGEVVASGSQDQTVRIWNVKTGE 1221
Score = 68.6 bits (166), Expect = 2e-11
Identities = 57/224 (25%), Positives = 101/224 (45%), Gaps = 3/224 (1%)
Frame = +1
Query: 133 IHASTKIDDTIRFLSLHDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKA 312
I AS D+ ++ S+ D I+ GH V +++ P + S S DKTI+LWD++
Sbjct: 698 ILASCGADENVKLWSVRDGVCIKTLTGHEHEVFSVAFHPDGETLASASGDKTIKLWDIQD 757
Query: 313 HNSQGFI--HTTGRPVGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDE 486
+ HT AF P+G + +++L+D+ +G + ++
Sbjct: 758 GTCLQTLTGHTDWVRCVAFSPDGNTLASSAADHTIKLWDV---SQGKCLR---TLKSHTG 811
Query: 487 W-NALKFSPNGRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYL 663
W ++ FS +G+ +A S IKI + G+ L ++ + +SPDS+ L
Sbjct: 812 WVRSVAFSADGQTLASGSGDRTIKIWNYHTGECLKTYI----GHTNSVYSIAYSPDSKIL 867
Query: 664 VCGAPDGQLCIWNTENKNRVALVAGFDHTPNSTSQPINCVAFNP 795
V G+ D + +W+ + + + G HT S VAF+P
Sbjct: 868 VSGSGDRTIKLWDCQTHICIKTLHG--HTNEVCS-----VAFSP 904
Score = 53.9 bits (128), Expect = 6e-07
Identities = 62/276 (22%), Positives = 108/276 (39%), Gaps = 6/276 (2%)
Frame = +1
Query: 97 NLMQFTHSSHTAIHASTKIDDTIRFLSLHDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGS 276
N++ S + A+ D +R + K + GH+ V + SP ++ S
Sbjct: 644 NILSAAFSPEGQLLATCDTDCHVRVWEVKSGKLLLICRGHSNWVRFVVFSPDGEILASCG 703
Query: 277 LDKTIRLWDLK----AHNSQGFIHTTGRPVGAFDPEGQIFVAGVNSESMRLYDIRNYDKG 444
D+ ++LW ++ G H AF P+G+ + ++++L+DI++
Sbjct: 704 ADENVKLWSVRDGVCIKTLTGHEHEVFSV--AFHPDGETLASASGDKTIKLWDIQD---- 757
Query: 445 PFVTFDVVQEAEDEWNALKFSPNGRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRV 624
+ D + FSP+G +A + IK+ D GK L L ++ V
Sbjct: 758 -GTCLQTLTGHTDWVRCVAFSPDGNTLASSAADHTIKLWDVSQGKCLRT---LKSHTGWV 813
Query: 625 PIDVCFSPDSQYLVCGAPDGQLCIWNTENKNRVALVAGFDHTPNSTSQPINCVAFNP--K 798
V FS D Q L G+ D + IWN + G HT + +A++P K
Sbjct: 814 R-SVAFSADGQTLASGSGDRTIKIWNYHTGECLKTYIG--HT-----NSVYSIAYSPDSK 865
Query: 799 YAMISSAVTNLTFWSPQIDE*IA*CKHIHVVKICFI 906
+ S + W Q I H H ++C +
Sbjct: 866 ILVSGSGDRTIKLWDCQTHICIK-TLHGHTNEVCSV 900
>sp|Q8YRI1|YY46_ANASP Hypothetical WD-repeat protein alr3466 Length = 1526 Score = 87.8 bits (216), Expect = 4e-17 Identities = 73/282 (25%), Positives = 127/282 (45%), Gaps = 5/282 (1%) Frame = +1 Query: 4 LISSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLH 183 L S S D + ++D + L VN + F T AS D T+R ++ Sbjct: 1173 LASGSGDQTVRLWDISSSKCLYILQGHTSWVNSVVFNPDGSTL--ASGSSDQTVRLWEIN 1230 Query: 184 DNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKA----HNSQGFIHTTGRP 351 +K + F GHT V ++ +P M SGS DKT+RLWD+ + H QG HT Sbjct: 1231 SSKCLCTFQGHTSWVNSVVFNPDGSMLASGSSDKTVRLWDISSSKCLHTFQG--HTNWVN 1288 Query: 352 VGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEW-NALKFSPNGRMIA 528 AF+P+G + +G +++RL++I + TF + W +++ FSP+G M+A Sbjct: 1289 SVAFNPDGSMLASGSGDQTVRLWEISS--SKCLHTF----QGHTSWVSSVTFSPDGTMLA 1342 Query: 529 ICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWNTE 708 S+ +++ G+ L F+ V FSPD L G+ D + +W+ Sbjct: 1343 SGSDDQTVRLWSISSGECLYTFL----GHTNWVGSVIFSPDGAILASGSGDQTVRLWSIS 1398 Query: 709 NKNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSAVTNLT 834 + + + G ++ + + F+P +++S + T Sbjct: 1399 SGKCLYTLQGHNNW-------VGSIVFSPDGTLLASGSDDQT 1433
Score = 76.6 bits (187), Expect = 9e-14
Identities = 70/244 (28%), Positives = 107/244 (43%), Gaps = 5/244 (2%)
Frame = +1
Query: 118 SSHTAIHASTKIDDTIRFLSLHDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRL 297
SS A+ AS D T+R + + GHT V ++ SP M SG D+ +RL
Sbjct: 1041 SSDGAMLASGSDDQTVRLWDISSGNCLYTLQGHTSCVRSVVFSPDGAMLASGGDDQIVRL 1100
Query: 298 WDLKAHNS----QGFIHTTGRPVGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDV 465
WD+ + N QG+ T+ F P G G + + +RL+DI + K T
Sbjct: 1101 WDISSGNCLYTLQGY--TSWVRFLVFSPNGVTLANGSSDQIVRLWDISS--KKCLYTL-- 1154
Query: 466 VQEAEDEW-NALKFSPNGRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCF 642
+ W NA+ FSP+G +A S +++ D K L IL + V V F
Sbjct: 1155 --QGHTNWVNAVAFSPDGATLASGSGDQTVRLWDISSSKCL---YILQGHTSWVN-SVVF 1208
Query: 643 SPDSQYLVCGAPDGQLCIWNTENKNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSAV 822
+PD L G+ D + +W + + G HT +N V FNP +M++S
Sbjct: 1209 NPDGSTLASGSSDQTVRLWEINSSKCLCTFQG--HT-----SWVNSVVFNPDGSMLASGS 1261
Query: 823 TNLT 834
++ T
Sbjct: 1262 SDKT 1265
Score = 75.1 bits (183), Expect = 3e-13
Identities = 57/205 (27%), Positives = 95/205 (46%), Gaps = 4/205 (1%)
Frame = +1
Query: 139 ASTKIDDTIRFLSLHDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKA-- 312
AS D T+R + + ++ F GHT RV ++ SP + M SGS D+T+RLWD+ +
Sbjct: 922 ASGSDDQTVRLWDISSGQCLKTFKGHTSRVRSVVFSPNSLMLASGSSDQTVRLWDISSGE 981
Query: 313 --HNSQGFIHTTGRPVGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDE 486
+ QG HT AF+ +G + G +++RL+DI + F + Q
Sbjct: 982 CLYIFQG--HTGWVYSVAFNLDGSMLATGSGDQTVRLWDISSSQ-----CFYIFQGHTSC 1034
Query: 487 WNALKFSPNGRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLV 666
++ FS +G M+A S+ +++ D G N + V FSPD L
Sbjct: 1035 VRSVVFSSDGAMLASGSDDQTVRLWDISSG----NCLYTLQGHTSCVRSVVFSPDGAMLA 1090
Query: 667 CGAPDGQLCIWNTENKNRVALVAGF 741
G D + +W+ + N + + G+
Sbjct: 1091 SGGDDQIVRLWDISSGNCLYTLQGY 1115
Score = 72.0 bits (175), Expect = 2e-12
Identities = 70/276 (25%), Positives = 129/276 (46%), Gaps = 5/276 (1%)
Frame = +1
Query: 4 LISSSYDDQMI-IYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSL 180
+++S DDQ + ++D +G + V + F S ++ + AS D T+R +
Sbjct: 920 MLASGSDDQTVRLWDISSGQCLKTFKGHTSRVRSVVF--SPNSLMLASGSSDQTVRLWDI 977
Query: 181 HDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFI----HTTGR 348
+ + F GHT V +++ + M +GS D+T+RLWD+ +SQ F HT+
Sbjct: 978 SSGECLYIFQGHTGWVYSVAFNLDGSMLATGSGDQTVRLWDIS--SSQCFYIFQGHTSCV 1035
Query: 349 PVGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRMIA 528
F +G + +G + +++RL+DI + + +Q ++ FSP+G M+A
Sbjct: 1036 RSVVFSSDGAMLASGSDDQTVRLWDISSGN-----CLYTLQGHTSCVRSVVFSPDGAMLA 1090
Query: 529 ICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWNTE 708
+G +I+ +D + L V + FSP+ L G+ D + +W+
Sbjct: 1091 ---SGGDDQIVRLWDISSGNCLYTLQGYTSWVRF-LVFSPNGVTLANGSSDQIVRLWDIS 1146
Query: 709 NKNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISS 816
+K + + G HT +N VAF+P A ++S
Sbjct: 1147 SKKCLYTLQG--HT-----NWVNAVAFSPDGATLAS 1175
Score = 56.2 bits (134), Expect = 1e-07
Identities = 55/220 (25%), Positives = 94/220 (42%), Gaps = 2/220 (0%)
Frame = +1
Query: 163 IRFLSLHDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFI--H 336
+RF K + GH V ++ S M SGS D+T+RLWD+ + H
Sbjct: 888 VRFWEAATGKELLTCKGHNSWVNSVGFSQDGKMLASGSDDQTVRLWDISSGQCLKTFKGH 947
Query: 337 TTGRPVGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNG 516
T+ F P + +G + +++RL+DI + + + Q ++ F+ +G
Sbjct: 948 TSRVRSVVFSPNSLMLASGSSDQTVRLWDISSGE-----CLYIFQGHTGWVYSVAFNLDG 1002
Query: 517 RMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCI 696
M+A GS + + +D F I + V V FS D L G+ D + +
Sbjct: 1003 SMLA---TGSGDQTVRLWDISSSQCFYIFQGHTSCVR-SVVFSSDGAMLASGSDDQTVRL 1058
Query: 697 WNTENKNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISS 816
W+ + N + + G HT + V F+P AM++S
Sbjct: 1059 WDISSGNCLYTLQG--HT-----SCVRSVVFSPDGAMLAS 1091
Score = 54.3 bits (129), Expect = 5e-07
Identities = 33/103 (32%), Positives = 55/103 (53%), Gaps = 3/103 (2%)
Frame = +1
Query: 130 AIHASTKIDDTIRFLSLHDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLK 309
AI AS D T+R S+ K + GH V ++ SP + SGS D+T+RLW++
Sbjct: 1381 AILASGSGDQTVRLWSISSGKCLYTLQGHNNWVGSIVFSPDGTLLASGSDDQTVRLWNIS 1440
Query: 310 AHNSQGFIH---TTGRPVGAFDPEGQIFVAGVNSESMRLYDIR 429
+ +H + R V AF +G I +G + E+++L+D++
Sbjct: 1441 SGECLYTLHGHINSVRSV-AFSSDGLILASGSDDETIKLWDVK 1482
Score = 35.0 bits (79), Expect = 0.30
Identities = 29/111 (26%), Positives = 50/111 (45%)
Frame = +1
Query: 502 FSPNGRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPD 681
FSP+G++ A +G ++ +A GK+L G N V FS D + L G+ D
Sbjct: 872 FSPDGKLFATGDSGGIVRFWEAATGKEL--LTCKGHNSW--VNSVGFSQDGKMLASGSDD 927
Query: 682 GQLCIWNTENKNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSAVTNLT 834
+ +W+ + + G HT + V F+P M++S ++ T
Sbjct: 928 QTVRLWDISSGQCLKTFKG--HTSR-----VRSVVFSPNSLMLASGSSDQT 971
>sp|P49695|PKWA_THECU Probable serine/threonine-protein kinase pkwA Length = 742 Score = 85.9 bits (211), Expect = 1e-16 Identities = 75/276 (27%), Positives = 126/276 (45%), Gaps = 3/276 (1%) Frame = +1 Query: 1 NLISSSYDDQMI-IYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLS 177 +L++ D++I ++D +G L V + F S A+ AS D T+R Sbjct: 472 SLLAGGSGDKLIHVWDVASGDELHTLEGHTDWVRAVAF--SPDGALLASGSDDATVRLWD 529 Query: 178 LHDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFI--HTTGRP 351 + + F GHT V+ ++ SP M SGS D T RLW++ + HT Sbjct: 530 VAAAEERAVFEGHTHYVLDIAFSPDGSMVASGSRDGTARLWNVATGTEHAVLKGHTDYVY 589 Query: 352 VGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRMIAI 531 AF P+G + +G ++RL+D+ + DV+Q + +L FSP+G M+ + Sbjct: 590 AVAFSPDGSMVASGSRDGTIRLWDVATGKER-----DVLQAPAENVVSLAFSPDGSML-V 643 Query: 532 CSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWNTEN 711 + S + + D G+ L F + V V FSPD L G+ D + +W+ Sbjct: 644 HGSDSTVHLWDVASGEALHTF---EGHTDWVRA-VAFSPDGALLASGSDDRTIRLWDVAA 699 Query: 712 KNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSA 819 + + G HT +P++ VAF+P+ ++SA Sbjct: 700 QEEHTTLEG--HT-----EPVHSVAFHPEGTTLASA 728
Score = 66.6 bits (161), Expect = 9e-11
Identities = 63/236 (26%), Positives = 102/236 (43%), Gaps = 4/236 (1%)
Frame = +1
Query: 4 LISSSYDDQMI-IYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSL 180
L++S DD + ++D + + V + S ++ AS D T R ++
Sbjct: 515 LLASGSDDATVRLWDVAAAEERAVFEGHTHYV--LDIAFSPDGSMVASGSRDGTARLWNV 572
Query: 181 HDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVG- 357
GHT V ++ SP M SGS D TIRLWD+ + + V
Sbjct: 573 ATGTEHAVLKGHTDYVYAVAFSPDGSMVASGSRDGTIRLWDVATGKERDVLQAPAENVVS 632
Query: 358 -AFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEW-NALKFSPNGRMIAI 531
AF P+G + V G +S ++ L+D+ + + TF E +W A+ FSP+G ++A
Sbjct: 633 LAFSPDGSMLVHGSDS-TVHLWDVASGEA--LHTF----EGHTDWVRAVAFSPDGALLA- 684
Query: 532 CSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIW 699
+GS + I +D L + + V V F P+ L + DG + IW
Sbjct: 685 --SGSDDRTIRLWDVAAQEEHTTLEGHTEPVH-SVAFHPEGTTLASASEDGTIRIW 737
Score = 60.8 bits (146), Expect = 5e-09
Identities = 40/141 (28%), Positives = 63/141 (44%), Gaps = 2/141 (1%)
Frame = +1
Query: 10 SSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLHDN 189
S S D + ++D TG + L + V + F+ +H S D T+ +
Sbjct: 602 SGSRDGTIRLWDVATGKERDVLQAPAENVVSLAFSPDGSMLVHGS---DSTVHLWDVASG 658
Query: 190 KYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVG--AF 363
+ + F GHT V ++ SP + SGS D+TIRLWD+ A + PV AF
Sbjct: 659 EALHTFEGHTDWVRAVAFSPDGALLASGSDDRTIRLWDVAAQEEHTTLEGHTEPVHSVAF 718
Query: 364 DPEGQIFVAGVNSESMRLYDI 426
PEG + ++R++ I
Sbjct: 719 HPEGTTLASASEDGTIRIWPI 739
Score = 30.4 bits (67), Expect = 7.4
Identities = 18/72 (25%), Positives = 33/72 (45%)
Frame = +1
Query: 619 RVPIDVCFSPDSQYLVCGAPDGQLCIWNTENKNRVALVAGFDHTPNSTSQPINCVAFNPK 798
R + V FSP L G+ D + +W+ + + + HT + + VAF+P
Sbjct: 460 REAVAVAFSPGGSLLAGGSGDKLIHVWDVASGDEL-------HTLEGHTDWVRAVAFSPD 512
Query: 799 YAMISSAVTNLT 834
A+++S + T
Sbjct: 513 GALLASGSDDAT 524
>sp|Q9V3J8|WDS_DROME Will die slowly protein Length = 361 Score = 85.9 bits (211), Expect = 1e-16 Identities = 66/275 (24%), Positives = 128/275 (46%), Gaps = 3/275 (1%) Frame = +1 Query: 4 LISSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLH 183 L SSS D + I+ G ++ ++ K G++ + ++ S + S D T++ L Sbjct: 87 LASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSRLLVSGSD--DKTLKVWELS 144 Query: 184 DNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVGA- 360 K ++ GH+ V + +P +++ +SGS D+++R+WD++ + PV A Sbjct: 145 TGKSLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVRTGKCLKTLPAHSDPVSAV 204 Query: 361 -FDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRMIAICS 537 F+ +G + V+ R++D + ++ + + +KFSPNG+ I + Sbjct: 205 HFNRDGSLIVSSSYDGLCRIWDTASGQ----CLKTLIDDDNPPVSFVKFSPNGKYILAAT 260 Query: 538 NGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFS-PDSQYLVCGAPDGQLCIWNTENK 714 + +K+ D GK L + G ++ I FS +++V G+ D + IWN ++K Sbjct: 261 LDNTLKLWDYSKGKCLKTY--TGHKNEKYCIFANFSVTGGKWIVSGSEDNMVYIWNLQSK 318 Query: 715 NRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSA 819 V + G T + C A +P +I+SA Sbjct: 319 EVVQKLQGHTDT-------VLCTACHPTENIIASA 346
Score = 53.9 bits (128), Expect = 6e-07
Identities = 52/228 (22%), Positives = 98/228 (42%), Gaps = 5/228 (2%)
Frame = +1
Query: 172 LSLHDNKYIRY-FVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFI--HTT 342
LS+ N +++ GHTK V + SP + S S DK I++W + I H
Sbjct: 56 LSVKPNYTLKFTLAGHTKAVSAVKFSPNGEWLASSSADKLIKIWGAYDGKFEKTISGHKL 115
Query: 343 GRPVGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRM 522
G A+ + ++ V+G + ++++++++ + ++ + F+P +
Sbjct: 116 GISDVAWSSDSRLLVSGSDDKTLKVWELSTGK-----SLKTLKGHSNYVFCCNFNPQSNL 170
Query: 523 IAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWN 702
I S ++I D GK L L A+ V V F+ D +V + DG IW+
Sbjct: 171 IVSGSFDESVRIWDVRTGKCLKT---LPAHSDPVSA-VHFNRDGSLIVSSSYDGLCRIWD 226
Query: 703 TENKNRVALVAGFDHTPNSTSQPINCVAFNP--KYAMISSAVTNLTFW 840
T + + + D + P++ V F+P KY + ++ L W
Sbjct: 227 TASGQCLKTLIDDD------NPPVSFVKFSPNGKYILAATLDNTLKLW 268
>sp|Q17963|YKY4_CAEEL Hypothetical WD-repeat protein C14B1.4 in chromosome III Length = 376 Score = 84.3 bits (207), Expect = 4e-16 Identities = 69/272 (25%), Positives = 127/272 (46%), Gaps = 3/272 (1%) Frame = +1 Query: 10 SSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLHDN 189 +SS D + I++ + +R L K GVN + ++ S + AS D T++ + + Sbjct: 104 TSSADKTVKIWNMDHMICERTLTGHKLGVNDIAWSSDSRCVVSASD--DKTLKIFEIVTS 161 Query: 190 KYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVGA--F 363 + + GH V + +P + + +SGS D+++R+WD+K + PV A F Sbjct: 162 RMTKTLKGHNNYVFCCNFNPQSSLVVSGSFDESVRIWDVKTGMCIKTLPAHSDPVSAVSF 221 Query: 364 DPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRMIAICSNG 543 + +G + +G +R++D N G + +V + +KFSPNG+ I + Sbjct: 222 NRDGSLIASGSYDGLVRIWDTAN---GQCIK-TLVDDENPPVAFVKFSPNGKYILASNLD 277 Query: 544 SQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFS-PDSQYLVCGAPDGQLCIWNTENKNR 720 S +K+ D GK L + G + I FS ++++ G+ D ++ IWN + + Sbjct: 278 STLKLWDFSKGKTLKQY--TGHENSKYCIFANFSVTGGKWIISGSEDCKIYIWNLQTREI 335 Query: 721 VALVAGFDHTPNSTSQPINCVAFNPKYAMISS 816 V + G HT QP+ +P +I+S Sbjct: 336 VQCLEG--HT-----QPVLASDCHPVQNIIAS 360
Score = 52.4 bits (124), Expect = 2e-06
Identities = 50/219 (22%), Positives = 95/219 (43%), Gaps = 9/219 (4%)
Frame = +1
Query: 211 GHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVG----AFDPEGQ 378
GHTK + + SP + S DKT+++W++ + + TG +G A+ + +
Sbjct: 85 GHTKSISSAKFSPCGKYLGTSSADKTVKIWNMDHMICERTL--TGHKLGVNDIAWSSDSR 142
Query: 379 IFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWN---ALKFSPNGRMIAICSNGSQ 549
V+ + +++++++I VT + + + N F+P ++ S
Sbjct: 143 CVVSASDDKTLKIFEI--------VTSRMTKTLKGHNNYVFCCNFNPQSSLVVSGSFDES 194
Query: 550 IKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWNTENKNRVAL 729
++I D G + L A+ V V F+ D + G+ DG + IW+T N +
Sbjct: 195 VRIWDVKTGMCIKT---LPAHSDPVSA-VSFNRDGSLIASGSYDGLVRIWDTANGQCIKT 250
Query: 730 VAGFDHTPNSTSQPINCVAFNP--KYAMISSAVTNLTFW 840
+ D P P+ V F+P KY + S+ + L W
Sbjct: 251 LVD-DENP-----PVAFVKFSPNGKYILASNLDSTLKLW 283
Score = 40.4 bits (93), Expect = 0.007
Identities = 26/101 (25%), Positives = 48/101 (47%), Gaps = 1/101 (0%)
Frame = +1
Query: 4 LISSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLH 183
++S S+D+ + I+D TG+ + L + V+ + F ++ AS D +R
Sbjct: 186 VVSGSFDESVRIWDVKTGMCIKTLPAHSDPVSAVSFNRDG--SLIASGSYDGLVRIWDTA 243
Query: 184 DNKYIRYFV-GHTKRVVTLSMSPVNDMFLSGSLDKTIRLWD 303
+ + I+ V V + SP L+ +LD T++LWD
Sbjct: 244 NGQCIKTLVDDENPPVAFVKFSPNGKYILASNLDSTLKLWD 284
>sp|P61964|WDR5_HUMAN WD-repeat protein 5 (BMP2-induced 3-kb gene protein) sp|P61965|WDR5_MOUSE WD-repeat protein 5 (BMP2-induced 3-kb gene protein) (WD-repeat protein BIG-3) Length = 334 Score = 83.6 bits (205), Expect = 7e-16 Identities = 62/257 (24%), Positives = 121/257 (47%), Gaps = 3/257 (1%) Frame = +1 Query: 4 LISSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLH 183 L SSS D + I+ G ++ ++ K G++ + ++ S+ + AS D T++ + Sbjct: 60 LASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSNLLVSASD--DKTLKIWDVS 117 Query: 184 DNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVGA- 360 K ++ GH+ V + +P +++ +SGS D+++R+WD+K + PV A Sbjct: 118 SGKCLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSAV 177 Query: 361 -FDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRMIAICS 537 F+ +G + V+ R++D + ++ + + +KFSPNG+ I + Sbjct: 178 HFNRDGSLIVSSSYDGLCRIWDTASGQ----CLKTLIDDDNPPVSFVKFSPNGKYILAAT 233 Query: 538 NGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFS-PDSQYLVCGAPDGQLCIWNTENK 714 + +K+ D GK L + G ++ I FS +++V G+ D + IWN + K Sbjct: 234 LDNTLKLWDYSKGKCLKTY--TGHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTK 291 Query: 715 NRVALVAGFDHTPNSTS 765 V + G ST+ Sbjct: 292 EIVQKLQGHTDVVISTA 308
Score = 52.0 bits (123), Expect = 2e-06
Identities = 49/214 (22%), Positives = 90/214 (42%), Gaps = 4/214 (1%)
Frame = +1
Query: 211 GHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFI--HTTGRPVGAFDPEGQIF 384
GHTK V ++ SP + S S DK I++W + I H G A+ + +
Sbjct: 43 GHTKAVSSVKFSPNGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSNLL 102
Query: 385 VAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRMIAICSNGSQIKIID 564
V+ + ++++++D+ + ++ + F+P +I S ++I D
Sbjct: 103 VSASDDKTLKIWDVSSGK-----CLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVRIWD 157
Query: 565 AFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWNTENKNRVALVAGFD 744
GK L L A+ V V F+ D +V + DG IW+T + + + D
Sbjct: 158 VKTGKCLKT---LPAHSDPVSA-VHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTLIDDD 213
Query: 745 HTPNSTSQPINCVAFNP--KYAMISSAVTNLTFW 840
+ P++ V F+P KY + ++ L W
Sbjct: 214 ------NPPVSFVKFSPNGKYILAATLDNTLKLW 241
>sp|Q00808|HETE1_PODAN Vegetatible incompatibility protein HET-E-1 Length = 1356 Score = 78.2 bits (191), Expect = 3e-14 Identities = 71/274 (25%), Positives = 117/274 (42%), Gaps = 4/274 (1%) Frame = +1 Query: 7 ISSSYDDQMI-IYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLH 183 ++S DD+ I I+D +G + L V + F+ AS D TI+ Sbjct: 898 VASGSDDKTIKIWDAASGTCTQTLEGHGGRVQSVAFSPDGQRV--ASGSDDHTIKIWDAA 955 Query: 184 DNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVG-- 357 + GH V++++ SP SGS DKTI++WD + + G V Sbjct: 956 SGTCTQTLEGHGSSVLSVAFSPDGQRVASGSGDKTIKIWDTASGTCTQTLEGHGGSVWSV 1015 Query: 358 AFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEW-NALKFSPNGRMIAIC 534 AF P+GQ +G + ++++++D + T E W ++ FSP+G+ +A Sbjct: 1016 AFSPDGQRVASGSDDKTIKIWDTAS------GTCTQTLEGHGGWVQSVVFSPDGQRVASG 1069 Query: 535 SNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWNTENK 714 S+ IKI DA G G + V FSPD Q + G+ DG + IW+ + Sbjct: 1070 SDDHTIKIWDAVSGTCTQTLEGHGDSVW----SVAFSPDGQRVASGSIDGTIKIWDAASG 1125 Query: 715 NRVALVAGFDHTPNSTSQPINCVAFNPKYAMISS 816 + G ++ VAF+P ++S Sbjct: 1126 TCTQTLEGHGGW-------VHSVAFSPDGQRVAS 1152
Score = 73.9 bits (180), Expect = 6e-13
Identities = 71/267 (26%), Positives = 110/267 (41%), Gaps = 19/267 (7%)
Frame = +1
Query: 7 ISSSYDDQMI-IYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLH 183
++S DD+ I I+D +G + L V + F+ AS D TI+
Sbjct: 1024 VASGSDDKTIKIWDTASGTCTQTLEGHGGWVQSVVFSPDGQRV--ASGSDDHTIKIWDAV 1081
Query: 184 DNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWD---------LKAHNSQGFIH 336
+ GH V +++ SP SGS+D TI++WD L+ H G++H
Sbjct: 1082 SGTCTQTLEGHGDSVWSVAFSPDGQRVASGSIDGTIKIWDAASGTCTQTLEGHG--GWVH 1139
Query: 337 TTGRPVGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEW-NALKFSPN 513
+ AF P+GQ +G +++++D + T E W ++ FSP+
Sbjct: 1140 SV-----AFSPDGQRVASGSIDGTIKIWDAASG------TCTQTLEGHGGWVQSVAFSPD 1188
Query: 514 GRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLC 693
G+ +A S+ IKI D G G Q V FSPD Q + G+ D +
Sbjct: 1189 GQRVASGSSDKTIKIWDTASGTCTQTLEGHGGWVQ----SVAFSPDGQRVASGSSDNTIK 1244
Query: 694 IWNTE--------NKNRVALVAGFDHT 750
IW+T N A FD+T
Sbjct: 1245 IWDTASGTCTQTLNVGSTATCLSFDYT 1271
Score = 64.7 bits (156), Expect = 4e-10
Identities = 53/208 (25%), Positives = 89/208 (42%), Gaps = 2/208 (0%)
Frame = +1
Query: 88 YGVNLMQFTHSSHTAIHASTKIDDTIRFLSLHDNKYIRYFVGHTKRVVTLSMSPVNDMFL 267
+G +++ S+ AS D TI+ + GH V +++ SP +
Sbjct: 840 HGSSVLSVAFSADGQRVASGSDDKTIKIWDTASGTGTQTLEGHGGSVWSVAFSPDRERVA 899
Query: 268 SGSLDKTIRLWDLKAHNSQGFIHTTGRPVG--AFDPEGQIFVAGVNSESMRLYDIRNYDK 441
SGS DKTI++WD + + G V AF P+GQ +G + +++++D +
Sbjct: 900 SGSDDKTIKIWDAASGTCTQTLEGHGGRVQSVAFSPDGQRVASGSDDHTIKIWDAAS--- 956
Query: 442 GPFVTFDVVQEAEDEWNALKFSPNGRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQR 621
++ ++ FSP+G+ +A S IKI D G G +
Sbjct: 957 --GTCTQTLEGHGSSVLSVAFSPDGQRVASGSGDKTIKIWDTASGTCTQTLEGHGGSVW- 1013
Query: 622 VPIDVCFSPDSQYLVCGAPDGQLCIWNT 705
V FSPD Q + G+ D + IW+T
Sbjct: 1014 ---SVAFSPDGQRVASGSDDKTIKIWDT 1038
Score = 61.6 bits (148), Expect = 3e-09
Identities = 54/227 (23%), Positives = 98/227 (43%), Gaps = 2/227 (0%)
Frame = +1
Query: 160 TIRFLSLHDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHT 339
TI + N + GH V++++ S SGS DKTI++WD + +
Sbjct: 822 TISVVEAEWNACTQTLEGHGSSVLSVAFSADGQRVASGSDDKTIKIWDTASGTGTQTLEG 881
Query: 340 TGRPVG--AFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPN 513
G V AF P+ + +G + ++++++D + ++ ++ FSP+
Sbjct: 882 HGGSVWSVAFSPDRERVASGSDDKTIKIWDAAS-----GTCTQTLEGHGGRVQSVAFSPD 936
Query: 514 GRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLC 693
G+ +A S+ IKI DA G G++ + V FSPD Q + G+ D +
Sbjct: 937 GQRVASGSDDHTIKIWDAASGTCTQTLEGHGSS----VLSVAFSPDGQRVASGSGDKTIK 992
Query: 694 IWNTENKNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSAVTNLT 834
IW+T + + G + + VAF+P ++S + T
Sbjct: 993 IWDTASGTCTQTLEGHGGS-------VWSVAFSPDGQRVASGSDDKT 1032
>sp|Q8YV57|Y2124_ANASP Hypothetical WD-repeat protein all2124 Length = 1683 Score = 76.6 bits (187), Expect = 9e-14 Identities = 72/252 (28%), Positives = 116/252 (46%), Gaps = 14/252 (5%) Frame = +1 Query: 1 NLISSSYDDQMI-IYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLS 177 +LI+S+ D+ + I+ G A + L VN + F+ T AS D+T++ + Sbjct: 1417 DLIASANADKTVKIWRVRDGKALKTLIGHDNEVNKVNFSPDGKTLASASR--DNTVKLWN 1474 Query: 178 LHDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLWD---------LKAHNSQGF 330 + D K+ + GHT V +S SP + S S DKTIRLWD L AHN Sbjct: 1475 VSDGKFKKTLKGHTDEVFWVSFSPDGKIIASASADKTIRLWDSFSGNLIKSLPAHND--L 1532 Query: 331 IHTTGRPVGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTF----DVVQEAEDEWNAL 498 +++ F+P+G + + ++++L+ R++D TF +VV + Sbjct: 1533 VYSVN-----FNPDGSMLASTSADKTVKLW--RSHDGHLLHTFSGHSNVVYSS------- 1578 Query: 499 KFSPNGRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAP 678 FSP+GR IA S +KI DG L+ Q + FSPD + L+ G+ Sbjct: 1579 SFSPDGRYIASASEDKTVKIWQ-IDGHLLTTL----PQHQAGVMSAIFSPDGKTLISGSL 1633 Query: 679 DGQLCIWNTENK 714 D IW +++ Sbjct: 1634 DTTTKIWRFDSQ 1645
Score = 65.9 bits (159), Expect = 2e-10
Identities = 62/257 (24%), Positives = 114/257 (44%), Gaps = 2/257 (0%)
Frame = +1
Query: 91 GVNLMQFTHSSHTAIHASTKIDDTIRFLSLHDNKYIRYFVGHTKRVVTLSMSPVNDMFLS 270
GV + F H +I A+ D I+ D ++ G+ K + +S +P D+ S
Sbjct: 1365 GVYAVSFLHDG--SIIATAGADGNIQLWHSQDGSLLKTLPGN-KAIYGISFTPQGDLIAS 1421
Query: 271 GSLDKTIRLWDLKAHNSQGFIHTTGRPVGA--FDPEGQIFVAGVNSESMRLYDIRNYDKG 444
+ DKT+++W ++ + + V F P+G+ + +++L+ N G
Sbjct: 1422 ANADKTVKIWRVRDGKALKTLIGHDNEVNKVNFSPDGKTLASASRDNTVKLW---NVSDG 1478
Query: 445 PFVTFDVVQEAEDEWNALKFSPNGRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRV 624
F ++ DE + FSP+G++IA S I++ D+F G + + L A+ V
Sbjct: 1479 KFK--KTLKGHTDEVFWVSFSPDGKIIASASADKTIRLWDSFSGNLIKS---LPAHNDLV 1533
Query: 625 PIDVCFSPDSQYLVCGAPDGQLCIWNTENKNRVALVAGFDHTPNSTSQPINCVAFNPKYA 804
V F+PD L + D + +W + + + + HT + S + +F+P
Sbjct: 1534 -YSVNFNPDGSMLASTSADKTVKLWRSHDGHLL-------HTFSGHSNVVYSSSFSPDGR 1585
Query: 805 MISSAVTNLTFWSPQID 855
I+SA + T QID
Sbjct: 1586 YIASASEDKTVKIWQID 1602
Score = 61.6 bits (148), Expect = 3e-09
Identities = 64/287 (22%), Positives = 115/287 (40%), Gaps = 9/287 (3%)
Frame = +1
Query: 1 NLISSSYDDQMIIYDCVTGVAKRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSL 180
NL S+S D + ++D +G L GV ++F+ T S D T++
Sbjct: 1169 NLASASSDHSIKLWDTTSGQLLMTLTGHSAGVITVRFSPDGQTIAAGSE--DKTVKLWHR 1226
Query: 181 HDNKYIRYFVGHTKRVVTLSMSPVNDMFLSGSLDKTIRLW---------DLKAHNSQGFI 333
D K ++ GH V +LS SP S S DKTI+LW LK HN +
Sbjct: 1227 QDGKLLKTLNGHQDWVNSLSFSPDGKTLASASADKTIKLWRIADGKLVKTLKGHNDSVWD 1286
Query: 334 HTTGRPVGAFDPEGQIFVAGVNSESMRLYDIRNYDKGPFVTFDVVQEAEDEWNALKFSPN 513
F +G+ + +++L++ + F A+ F P+
Sbjct: 1287 VN-------FSSDGKAIASASRDNTIKLWNRHGIELETFTGH------SGGVYAVNFLPD 1333
Query: 514 GRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCGAPDGQLC 693
+IA S + I++ + +S +L N V F D + DG +
Sbjct: 1334 SNIIASASLDNTIRL---WQRPLISPLEVLAGNSG--VYAVSFLHDGSIIATAGADGNIQ 1388
Query: 694 IWNTENKNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSAVTNLT 834
+W++++ + + + G ++ I ++F P+ +I+SA + T
Sbjct: 1389 LWHSQDGSLLKTLPG--------NKAIYGISFTPQGDLIASANADKT 1427
Score = 60.8 bits (146), Expect = 5e-09
Identities = 59/227 (25%), Positives = 92/227 (40%), Gaps = 2/227 (0%)
Frame = +1
Query: 64 KRHLNSKKYGVNLMQFTHSSHTAIHASTKIDDTIRFLSLHDNKYIRYFVGHTKRVVTLSM 243
+ L K GV + + T AS +D TI+ S D + R GH V ++S
Sbjct: 1065 RNRLEGHKDGVISISISRDGQTI--ASGSLDKTIKLWS-RDGRLFRTLNGHEDAVYSVSF 1121
Query: 244 SPVNDMFLSGSLDKTIRLWDLKAHNSQGFIHTTGRPVG--AFDPEGQIFVAGVNSESMRL 417
SP SG DKTI+LW I + V F P+G+ + + S++L
Sbjct: 1122 SPDGQTIASGGSDKTIKLWQTSDGTLLKTITGHEQTVNNVYFSPDGKNLASASSDHSIKL 1181
Query: 418 YDIRNYDKGPFVTFDVVQEAEDEWNALKFSPNGRMIAICSNGSQIKIIDAFDGKQLSNFM 597
+D + +T ++FSP+G+ IA S +K+ DGK L
Sbjct: 1182 WDTTSGQLLMTLTGHSAGVI-----TVRFSPDGQTIAAGSEDKTVKLWHRQDGKLLKT-- 1234
Query: 598 ILGANPQRVPIDVCFSPDSQYLVCGAPDGQLCIWNTENKNRVALVAG 738
L + V + FSPD + L + D + +W + V + G
Sbjct: 1235 -LNGHQDWVN-SLSFSPDGKTLASASADKTIKLWRIADGKLVKTLKG 1279
Score = 35.0 bits (79), Expect = 0.30
Identities = 30/109 (27%), Positives = 48/109 (44%)
Frame = +1
Query: 493 ALKFSPNGRMIAICSNGSQIKIIDAFDGKQLSNFMILGANPQRVPIDVCFSPDSQYLVCG 672
++ S +G+ IA S IK+ DG+ F L + V V FSPD Q + G
Sbjct: 1077 SISISRDGQTIASGSLDKTIKLWSR-DGRL---FRTLNGHEDAV-YSVSFSPDGQTIASG 1131
Query: 673 APDGQLCIWNTENKNRVALVAGFDHTPNSTSQPINCVAFNPKYAMISSA 819
D + +W T + + + G + T +N V F+P ++SA
Sbjct: 1132 GSDKTIKLWQTSDGTLLKTITGHEQT-------VNNVYFSPDGKNLASA 1173
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 108,681,305
Number of Sequences: 369166
Number of extensions: 2284674
Number of successful extensions: 8062
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5835
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7153
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 9606826000
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail