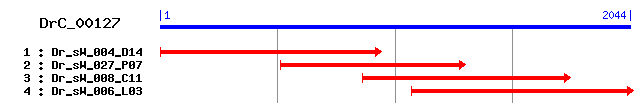
DrC_00127
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00127 (2044 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q10580|SWAP_CAEEL SWAP protein (Suppressor of white apri... 82 5e-15 sp|Q12872|SFRS8_HUMAN Splicing factor, arginine/serine-rich... 77 2e-13 sp|P12297|SUWA_DROME Suppressor of white apricot protein 76 3e-13 sp|Q8K4Z5|SF3A1_MOUSE Splicing factor 3 subunit 1 (SF3a120) 52 9e-06 sp|Q15459|SF3A1_HUMAN Splicing factor 3 subunit 1 (Spliceos... 51 1e-05 sp|Q8CFC7|SFR16_MOUSE Splicing factor, arginine/serine-rich... 47 3e-04 sp|Q8N2M8|SFR16_HUMAN Splicing factor, arginine/serine-rich... 47 3e-04 sp|Q68FU8|SF04_RAT Splicing factor 4 40 0.034 sp|Q8CH02|SF04_MOUSE Splicing factor 4 39 0.057 sp|Q8IWZ8|SF04_HUMAN Splicing factor 4 (RNA-binding protein... 39 0.057
>sp|Q10580|SWAP_CAEEL SWAP protein (Suppressor of white apricot protein homolog) Length = 749 Score = 82.4 bits (202), Expect = 5e-15 Identities = 49/140 (35%), Positives = 76/140 (54%), Gaps = 1/140 (0%) Frame = +3 Query: 435 IDLQSACESPGNEDEYNSKRYDQSRYSAIHFDYDNNQEKNESQESKYEKEVEDNGTDAAY 614 +D+ + ++E +R DQ +AI FDY + K +S+ D + Sbjct: 96 LDMYKDIQREQEKEEEEKRRNDQR--NAIGFDYGTGKVKARESDSE----------DEPF 143 Query: 615 LCPPNLKLLPGMITPATEREAAIIEQTAKFVADKGPQLEILIKAKQSNN-PKFVFLNFDH 791 P +K G+ P+ + IIE+TA F+ G Q+EI+IKAKQ NN +F FL FDH Sbjct: 144 EPPEGIKFPVGLELPSNMKLHHIIEKTASFIVANGTQMEIVIKAKQRNNAEQFGFLEFDH 203 Query: 792 KLYPFYRHVTQLIKTAQYIP 851 +L PFY+++ +LI+ +YIP Sbjct: 204 RLNPFYKYLQKLIREKKYIP 223
Score = 38.5 bits (88), Expect = 0.075
Identities = 25/77 (32%), Positives = 36/77 (46%), Gaps = 4/77 (5%)
Frame = +3
Query: 9 TLFQDDIKSKEINRGALLVPYNGDKSIMIDRFDCRGHLND----LQSHGGPFNELQLSHE 176
T+F +D +S+ I VP GD +DR+DCR L ++ +G P +
Sbjct: 26 TIFPNDYQSEHIAEERHTVPCLGDPENRVDRYDCRLLLPSIDVAIKRNGSPSEQCPTEAM 85
Query: 177 EWEVEQSCNFERYNDLY 227
E E C ERY D+Y
Sbjct: 86 E---EDMCEEERYLDMY 99
Score = 34.3 bits (77), Expect = 1.4
Identities = 42/164 (25%), Positives = 70/164 (42%), Gaps = 10/164 (6%)
Frame = +3
Query: 549 KNESQESKYEKEVEDNGTDAAYL----------CPPNLKLLPGMITPATEREAAIIEQTA 698
K +E+K EKE D+ D Y CP + P + PAT I+ A
Sbjct: 341 KMNVEEAKKEKE-NDHLDDPEYREWYENFYGRPCP---WIGPRPMIPATPDLEPILNSYA 396
Query: 699 KFVADKGPQLEILIKAKQSNNPKFVFLNFDHKLYPFYRHVTQLIKTAQYIPGKRHISHLV 878
+ VA +G + E + A++ + + F+ Y +Y H ++ + Y P ++++S LV
Sbjct: 397 EHVAQRGLEAEASLAARE--DLQLHFMEPKSPYYSYYHHKVRMHQWRMYQPIEQNLSPLV 454
Query: 879 ESNGDQSQMGSFPQPVAPKIDISSTPYGKLIERFKQSQEKKKLD 1010
++ S P P + STP L R Q ++ LD
Sbjct: 455 LNSPAPPSAVSSPGPSSLMSLNLSTPEPPLNRR----QRRRLLD 494
>sp|Q12872|SFRS8_HUMAN Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog) Length = 951 Score = 77.0 bits (188), Expect = 2e-13 Identities = 49/145 (33%), Positives = 79/145 (54%), Gaps = 15/145 (10%) Frame = +3 Query: 456 ESPGNEDEYNSKRYDQS-----RYSAIHF----DYDNNQEKNESQESKYEKE------VE 590 E E+EY KR+ ++ Y+A+ F DY + E E +E ++E +E Sbjct: 125 EEARQEEEY--KRFSEALAEDGSYNAVGFTYGSDYYDPSEPTEEEEPSKQREKNEAENLE 182 Query: 591 DNGTDAAYLCPPNLKLLPGMITPATEREAAIIEQTAKFVADKGPQLEILIKAKQSNNPKF 770 +N + ++ P L + + P T + AIIE+TA FV +G Q EI++KAKQ+ N +F Sbjct: 183 EN--EEPFVAPLGLSVPSDVELPPTAKMHAIIERTASFVCRQGAQFEIMLKAKQAPNSQF 240 Query: 771 VFLNFDHKLYPFYRHVTQLIKTAQY 845 FL FDH L P+Y+ + + +K +Y Sbjct: 241 DFLRFDHYLNPYYKFIQKAMKEGRY 265
Score = 65.5 bits (158), Expect = 6e-10
Identities = 32/79 (40%), Positives = 50/79 (63%), Gaps = 4/79 (5%)
Frame = +3
Query: 3 SCTLFQDDIKSKEINRGALLVPYNGDKSIMIDRFDCRGHLNDLQSHGGPFN----ELQLS 170
+C LF+DD +++ +G L+P+ GD I+IDR+D RGHL+DL + ++ + QLS
Sbjct: 41 ACKLFRDDERAQAQEQGQHLIPWMGDHKILIDRYDGRGHLHDLSEYDAEYSTWNRDYQLS 100
Query: 171 HEEWEVEQSCNFERYNDLY 227
EE +E C+ ERY L+
Sbjct: 101 EEEARIEALCDEERYLALH 119
Score = 49.7 bits (117), Expect = 3e-05
Identities = 25/65 (38%), Positives = 39/65 (60%)
Frame = +3
Query: 1374 VIGKMAEYVARNGVEFEQMMLARNSERFAFLLPGHASHCRYKILLDKELNKQQQQTIGRS 1553
VI K+AEYVARNG++FE + A+N +RF FL P + + Y+ L K+ ++
Sbjct: 459 VIDKLAEYVARNGLKFETSVRAKNDQRFEFLQPWYQYNAYYEFKKQFFLQKEGGDSMQAV 518
Query: 1554 QSPEQ 1568
+PE+
Sbjct: 519 SAPEE 523
>sp|P12297|SUWA_DROME Suppressor of white apricot protein Length = 963 Score = 76.3 bits (186), Expect = 3e-13 Identities = 45/120 (37%), Positives = 68/120 (56%), Gaps = 6/120 (5%) Frame = +3 Query: 510 YSAIHFDYDNNQEKNES--QESKYEKEVEDNGTDAA--YLCPPNLKLLP--GMITPATER 671 +S + F YD + S S ++ N ++ ++ P L + P M P T + Sbjct: 171 FSQVGFQYDGQSAASTSIGGSSTATSQLSPNSEESELPFVLPYTLMMAPPLDMQLPETMK 230 Query: 672 EAAIIEQTAKFVADKGPQLEILIKAKQSNNPKFVFLNFDHKLYPFYRHVTQLIKTAQYIP 851 + AIIE+TA+F+A +G Q+EILIKAKQ+NN +F FL L P+YRH+ IK A++ P Sbjct: 231 QHAIIEKTARFIATQGAQMEILIKAKQANNTQFDFLTQGGHLQPYYRHLLAAIKAAKFPP 290
Score = 66.2 bits (160), Expect = 3e-10
Identities = 36/84 (42%), Positives = 52/84 (61%), Gaps = 5/84 (5%)
Frame = +3
Query: 3 SCTLFQDDIKSKEINRGALLVPYNGDKSIMIDRFDCRGHLNDLQSH----GGPFNELQ-L 167
+C +F+DD K++E++ G L+P+ GD ++ IDR+D RG L +L H GG N L+ L
Sbjct: 66 ACKIFRDDEKAREMDHGKQLIPWMGDVNLKIDRYDVRGALCELAPHEAPPGGYGNRLEYL 125
Query: 168 SHEEWEVEQSCNFERYNDLYHTND 239
S EE EQ C ERY LY+ +
Sbjct: 126 SAEEQRAEQLCEEERYLFLYNNEE 149
Score = 41.2 bits (95), Expect = 0.012
Identities = 47/219 (21%), Positives = 91/219 (41%), Gaps = 37/219 (16%)
Frame = +3
Query: 1356 EESQDAVIGKMAEYVARNGVEFEQMMLARNSERFAFLLPG--HASHCRYKILLDKELNKQ 1529
++S +I K A YV +NG +FE+ + ++ +RF+FLLP + + YK+ D + +
Sbjct: 477 KDSLRHIIDKTATYVIKNGRQFEETLRTKSVDRFSFLLPANEYYPYYLYKVTGDVDAASK 536
Query: 1530 QQQTIGRSQSPEQCIASIVDSYG-----QSPPHQSSGPISFKVTSK------LRTVPVRS 1676
+++T + ++ S+G S + P+SF + ++ T+P +
Sbjct: 537 EEKTRKAAAVAAALMSKKGLSFGGAAAAVSGSNLDKAPVSFSIRARDDQCPLQHTLPQEA 596
Query: 1677 SN--YVDTIAQIEXXXXXXXXXXNLQVKSEITNSSSRTT-KNGRIS-------------- 1805
S+ A +E +K T +RT + G I+
Sbjct: 597 SDEETSSNAAGVEHVRPGMPDSVQRAIKQVETQLLARTAGQKGNITASPSCSSPQKEQRQ 656
Query: 1806 -------RFSPANPKNLMVLATFKNDLQIQRKRKAAEFL 1901
+ + + L + + + LQ++RKRKA FL
Sbjct: 657 AEERVKDKLAQIAREKLNGMISREKQLQLERKRKALAFL 695
>sp|Q8K4Z5|SF3A1_MOUSE Splicing factor 3 subunit 1 (SF3a120) Length = 791 Score = 51.6 bits (122), Expect = 9e-06 Identities = 33/106 (31%), Positives = 51/106 (48%) Frame = +3 Query: 621 PPNLKLLPGMITPATEREAAIIEQTAKFVADKGPQLEILIKAKQSNNPKFVFLNFDHKLY 800 P K + G+I P E I+++TA FVA GP+ E I+ + NNPKF FLN + + Sbjct: 33 PTPSKPVVGIIYPPPEVRN-IVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPNDPYH 91 Query: 801 PFYRHVTQLIKTAQYIPGKRHISHLVESNGDQSQMGSFPQPVAPKI 938 +YRH K + I +++ Q+ PQ V ++ Sbjct: 92 AYYRHKVSEFKEGKAQEPSAAIPKVMQQQ-QQATQQQLPQKVQAQV 136
Score = 38.9 bits (89), Expect = 0.057
Identities = 24/83 (28%), Positives = 38/83 (45%), Gaps = 2/83 (2%)
Frame = +3
Query: 1374 VIGKMAEYVARNGVEFEQMMLAR--NSERFAFLLPGHASHCRYKILLDKELNKQQQQTIG 1547
++ K A +VARNG EFE + N+ +F FL P H Y+ +K + G
Sbjct: 52 IVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPNDPYHAYYR-------HKVSEFKEG 104
Query: 1548 RSQSPEQCIASIVDSYGQSPPHQ 1616
++Q P I ++ Q+ Q
Sbjct: 105 KAQEPSAAIPKVMQQQQQATQQQ 127
Score = 33.1 bits (74), Expect = 3.1
Identities = 29/110 (26%), Positives = 52/110 (47%), Gaps = 2/110 (1%)
Frame = +3
Query: 681 IIEQTAKFVADKGPQLEILIKAKQSNNPKFVFLNFDHKLYPFYRHVTQLIK--TAQYIPG 854
+++ TA+FVA G Q + K+ N +F FL H L+ ++ T+L++ T IP
Sbjct: 166 VVKLTAQFVARNGRQFLTQLMQKEQRNYQFDFLRPQHSLFNYF---TKLVEQYTKILIPP 222
Query: 855 KRHISHLVESNGDQSQMGSFPQPVAPKIDISSTPYGKLIERFKQSQEKKK 1004
K S L + P+ V ++ + K ER ++ +E++K
Sbjct: 223 KGLFSKL-------KKEAENpreVLDQV-CYRVEWAKFQERERKKEEEEK 264
>sp|Q15459|SF3A1_HUMAN Splicing factor 3 subunit 1 (Spliceosome associated protein 114) (SAP 114) (SF3a120) Length = 793 Score = 50.8 bits (120), Expect = 1e-05 Identities = 32/102 (31%), Positives = 50/102 (49%) Frame = +3 Query: 633 KLLPGMITPATEREAAIIEQTAKFVADKGPQLEILIKAKQSNNPKFVFLNFDHKLYPFYR 812 K + G+I P E I+++TA FVA GP+ E I+ + NNPKF FLN + + +YR Sbjct: 37 KPVVGIIYPPPEVRN-IVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPNDPYHAYYR 95 Query: 813 HVTQLIKTAQYIPGKRHISHLVESNGDQSQMGSFPQPVAPKI 938 H K + I +++ Q+ PQ V ++ Sbjct: 96 HKVSEFKEGKAQEPSAAIPKVMQQQ-QQTTQQQLPQKVQAQV 136
Score = 38.9 bits (89), Expect = 0.057
Identities = 24/83 (28%), Positives = 38/83 (45%), Gaps = 2/83 (2%)
Frame = +3
Query: 1374 VIGKMAEYVARNGVEFEQMMLAR--NSERFAFLLPGHASHCRYKILLDKELNKQQQQTIG 1547
++ K A +VARNG EFE + N+ +F FL P H Y+ +K + G
Sbjct: 52 IVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPNDPYHAYYR-------HKVSEFKEG 104
Query: 1548 RSQSPEQCIASIVDSYGQSPPHQ 1616
++Q P I ++ Q+ Q
Sbjct: 105 KAQEPSAAIPKVMQQQQQTTQQQ 127
Score = 33.1 bits (74), Expect = 3.1
Identities = 29/110 (26%), Positives = 52/110 (47%), Gaps = 2/110 (1%)
Frame = +3
Query: 681 IIEQTAKFVADKGPQLEILIKAKQSNNPKFVFLNFDHKLYPFYRHVTQLIK--TAQYIPG 854
+++ TA+FVA G Q + K+ N +F FL H L+ ++ T+L++ T IP
Sbjct: 166 VVKLTAQFVARNGRQFLTQLMQKEQRNYQFDFLRPQHSLFNYF---TKLVEQYTKILIPP 222
Query: 855 KRHISHLVESNGDQSQMGSFPQPVAPKIDISSTPYGKLIERFKQSQEKKK 1004
K S L + P+ V ++ + K ER ++ +E++K
Sbjct: 223 KGLFSKL-------KKEAENpreVLDQV-CYRVEWAKFQERERKKEEEEK 264
>sp|Q8CFC7|SFR16_MOUSE Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein) Length = 653 Score = 46.6 bits (109), Expect = 3e-04 Identities = 25/82 (30%), Positives = 41/82 (50%) Frame = +3 Query: 3 SCTLFQDDIKSKEINRGALLVPYNGDKSIMIDRFDCRGHLNDLQSHGGPFNELQLSHEEW 182 +C + D + ++P+ GD + MIDRFD R HL+ + + P +S E+ Sbjct: 32 ACKVHLDSAVALAAESPVNMMPWQGDTNNMIDRFDVRAHLDHIPDYTPPL-LTTISPEQE 90 Query: 183 EVEQSCNFERYNDLYHTNDSDI 248 E+ CN+ERY L + + I Sbjct: 91 SDERKCNYERYRGLVQNDFAGI 112
>sp|Q8N2M8|SFR16_HUMAN Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) Length = 659 Score = 46.6 bits (109), Expect = 3e-04 Identities = 25/82 (30%), Positives = 41/82 (50%) Frame = +3 Query: 3 SCTLFQDDIKSKEINRGALLVPYNGDKSIMIDRFDCRGHLNDLQSHGGPFNELQLSHEEW 182 +C + D + ++P+ GD + MIDRFD R HL+ + + P +S E+ Sbjct: 32 ACKVHLDSAVALAAESPVNMMPWQGDTNNMIDRFDVRAHLDHIPDYTPPL-LTTISPEQE 90 Query: 183 EVEQSCNFERYNDLYHTNDSDI 248 E+ CN+ERY L + + I Sbjct: 91 SDERKCNYERYRGLVQNDFAGI 112
>sp|Q68FU8|SF04_RAT Splicing factor 4 Length = 644 Score = 39.7 bits (91), Expect = 0.034 Identities = 39/161 (24%), Positives = 66/161 (40%), Gaps = 18/161 (11%) Frame = +3 Query: 528 DYDNN------QEKNESQESKYEKEVEDNGTDAAYLCPPNLKLLPGMITPATEREAAIIE 689 DY +N +KN + Y K+V + +A K+ P P E + E Sbjct: 210 DYKDNPAFTFLHDKNSREFLYYRKKVAEIRKEAQKPQAATQKVSP----PEDEEAKNLAE 265 Query: 690 QTAKFVADKGPQLEILIKAKQSNNPKFVFL-NFDHKLYPFYRHVTQLIKTAQY------- 845 + A+F+AD GP++E + N F FL + + + Y +Y+ + + A+ Sbjct: 266 KLARFIADGGPEVETIALQNNRENQAFSFLYDPNSQGYRYYKQKLEEFRKAKAGSTGSLP 325 Query: 846 ----IPGKRHISHLVESNGDQSQMGSFPQPVAPKIDISSTP 956 P R S +G + + P PVAP ++ TP Sbjct: 326 APVPNPSLRRKSAPEALSGAVPPITACPTPVAPAPAVNPTP 366
Score = 36.6 bits (83), Expect = 0.28
Identities = 21/56 (37%), Positives = 35/56 (62%), Gaps = 2/56 (3%)
Frame = +3
Query: 681 IIEQTAKFVADKGPQLEILIKAKQSNNPKFVFL-NFDHKLYPFYR-HVTQLIKTAQ 842
+IE+ A+FVA+ GP+LE + +NP F FL + + + + +YR V ++ K AQ
Sbjct: 188 VIEKLARFVAEGGPELEKVAMEDYKDNPAFTFLHDKNSREFLYYRKKVAEIRKEAQ 243
>sp|Q8CH02|SF04_MOUSE Splicing factor 4 Length = 643 Score = 38.9 bits (89), Expect = 0.057 Identities = 39/161 (24%), Positives = 65/161 (40%), Gaps = 18/161 (11%) Frame = +3 Query: 528 DYDNN------QEKNESQESKYEKEVEDNGTDAAYLCPPNLKLLPGMITPATEREAAIIE 689 DY +N +KN + Y ++V + +A K+ P P E + E Sbjct: 209 DYKDNPAFTFLHDKNSREFLYYRRKVAEIRKEAQKPQAATQKVSP----PEDEEAKNLAE 264 Query: 690 QTAKFVADKGPQLEILIKAKQSNNPKFVFL-NFDHKLYPFYRHVTQLIKTAQY------- 845 + A+F+AD GP++E + N F FL + + + Y +YR + A+ Sbjct: 265 KLARFIADGGPEVETIALQNNRENQAFSFLYDPNSQGYRYYRQKLDEFRKAKAGSTGSFP 324 Query: 846 ----IPGKRHISHLVESNGDQSQMGSFPQPVAPKIDISSTP 956 P R S +G + + P PVAP ++ TP Sbjct: 325 APAPNPSLRRKSAPEALSGAVPPITACPTPVAPAPAVNPTP 365
Score = 38.5 bits (88), Expect = 0.075
Identities = 31/103 (30%), Positives = 48/103 (46%), Gaps = 3/103 (2%)
Frame = +3
Query: 543 QEKNESQESKYEKEVEDNGTDAAYLCPPNLKLLPGMITPATEREAAIIEQTAKFVADKGP 722
Q ++ +E YE+ +E +K+ P P +IE+ A+FVA+ GP
Sbjct: 158 QSPDDDEEEDYEQWLE-------------IKVSP----PEGAETRRVIEKLARFVAEGGP 200
Query: 723 QLEILIKAKQSNNPKFVFL---NFDHKLYPFYRHVTQLIKTAQ 842
+LE + +NP F FL N LY + R V ++ K AQ
Sbjct: 201 ELEKVAMEDYKDNPAFTFLHDKNSREFLY-YRRKVAEIRKEAQ 242
>sp|Q8IWZ8|SF04_HUMAN Splicing factor 4 (RNA-binding protein RBP) Length = 645 Score = 38.9 bits (89), Expect = 0.057 Identities = 30/102 (29%), Positives = 50/102 (49%), Gaps = 2/102 (1%) Frame = +3 Query: 543 QEKNESQESKYEKEVEDNGTDAAYLCPPNLKLLPGMITPATEREAAIIEQTAKFVADKGP 722 Q +E +E YE+ +E +K+ P P +IE+ A+FVA+ GP Sbjct: 162 QSPDEDEEEDYEQWLE-------------IKVSP----PEGAETRKVIEKLARFVAEGGP 204 Query: 723 QLEILIKAKQSNNPKFVFL-NFDHKLYPFYR-HVTQLIKTAQ 842 +LE + +NP F FL + + + + +YR V ++ K AQ Sbjct: 205 ELEKVAMEDYKDNPAFAFLHDKNSREFLYYRKKVAEIRKEAQ 246
Score = 37.4 bits (85), Expect = 0.17
Identities = 29/112 (25%), Positives = 50/112 (44%), Gaps = 7/112 (6%)
Frame = +3
Query: 528 DYDNN------QEKNESQESKYEKEVEDNGTDAAYLCPPNLKLLPGMITPATEREAAIIE 689
DY +N +KN + Y K+V + +A + K+ P P E + E
Sbjct: 213 DYKDNPAFAFLHDKNSREFLYYRKKVAEIRKEAQKSQAASQKVSP----PEDEEVKNLAE 268
Query: 690 QTAKFVADKGPQLEILIKAKQSNNPKFVFL-NFDHKLYPFYRHVTQLIKTAQ 842
+ A+F+AD GP++E + N F FL + + Y +YR + + A+
Sbjct: 269 KLARFIADGGPEVETIALQNNRENQAFSFLYEPNSQGYKYYRQKLEEFRKAK 320
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 215,708,435
Number of Sequences: 369166
Number of extensions: 4207611
Number of successful extensions: 11417
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 10635
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11404
length of database: 68,354,980
effective HSP length: 117
effective length of database: 46,740,985
effective search space used: 26315174555
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail