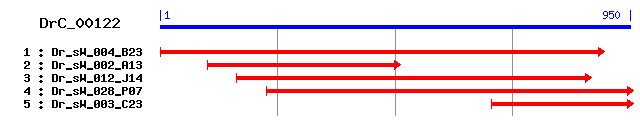
DrC_00122
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00122 (950 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P24733|MYS_AEQIR Myosin heavy chain, striated muscle 278 1e-74 sp|P05661|MYSA_DROME Myosin heavy chain, muscle 278 1e-74 sp|P02566|MYO4_CAEEL Myosin-4 (Myosin heavy chain B) (MHC B... 251 3e-66 sp|P12844|MYO3_CAEEL Myosin-3 (Myosin heavy chain A) (MHC A) 236 7e-62 sp|P02565|MYH3_CHICK Myosin heavy chain, fast skeletal musc... 232 1e-60 sp|P13538|MYSS_CHICK Myosin heavy chain, skeletal muscle, a... 231 3e-60 sp|Q5SX40|MYH1_MOUSE Myosin-1 (Myosin heavy chain, skeletal... 229 9e-60 sp|P13533|MYH6_HUMAN Myosin heavy chain, cardiac muscle alp... 229 9e-60 sp|P13539|MYH6_MESAU Myosin heavy chain, cardiac muscle alp... 229 1e-59 sp|Q9UKX2|MYH2_HUMAN Myosin heavy chain, skeletal muscle, a... 228 2e-59
>sp|P24733|MYS_AEQIR Myosin heavy chain, striated muscle Length = 1938 Score = 278 bits (712), Expect = 1e-74 Identities = 139/245 (56%), Positives = 185/245 (75%) Frame = +2 Query: 2 VRGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNE 181 +R +LE +ER RK +++E + DR+NEL+ Q SS KRKLE D+ AMQ+DL+E E Sbjct: 1685 LRAALEQAERARKASDNELADANDRVNELTSQVSSVQGQKRKLEGDINAMQTDLDEMHGE 1744 Query: 182 ARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXX 361 + A+E+ KKA+AD++RL DE+R EQ+H+ Q++K++K LE+Q KE Q +LDE+E +++ Sbjct: 1745 LKGADERCKKAMADAARLADELRAEQDHSNQVEKVRKNLESQVKEFQIRLDEAEASSLKG 1804 Query: 362 XXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDL 541 E RV ELE+ELD EQ+RH ETQKN RK +RRLKE+ Q DED+KNQERLQ+L Sbjct: 1805 GKKMIQKLESRVHELEAELDNEQRRHAETQKNMRKADRRLKELAFQADEDRKNQERLQEL 1864 Query: 542 VEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSS 721 ++KL KIKT+KRQVEEAEEIAA+NLAKYRK Q E+E++EERAD A+ LQK R K+RSS Sbjct: 1865 IDKLNAKIKTFKRQVEEAEEIAAINLAKYRKAQHELEEAEERADTADSTLQKFRAKSRSS 1924 Query: 722 VSTAR 736 VS R Sbjct: 1925 VSVQR 1929
Score = 89.7 bits (221), Expect = 1e-17
Identities = 59/227 (25%), Positives = 114/227 (50%)
Frame = +2
Query: 47 ESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQAKKAVADS 226
ES+ EL +RL + ++ K+K+EAD A ++ D+ + N ++A +
Sbjct: 913 ESQIKELEERLLDEEDAAADLEGIKKKMEADNANLKKDIGDLENTLQKAEQDKAHKDNQI 972
Query: 227 SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXXEQRVREL 406
S L EI Q+ EH +++K KK LE NK+ L ++E + EQ + EL
Sbjct: 973 STLQGEISQQDEHIGKLNKEKKALEEANKKTSDSL-QAEEDKCNHLNKLKAKLEQALDEL 1031
Query: 407 ESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQV 586
E L+ E+K + +K RK+E+ LK ++ ++ + L++ V + + +I + ++
Sbjct: 1032 EDNLEREKKVRGDVEKAKRKVEQDLKSTQENVEDLERVKRELEENVRRKEAEISSLNSKL 1091
Query: 587 EEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVS 727
E+ + + + K +++Q IE+ EE + A K+ K R+ ++
Sbjct: 1092 EDEQNLVSQLQRKIKELQARIEELEEELEAERNARAKVE-KQRAELN 1137
Score = 80.9 bits (198), Expect = 5e-15
Identities = 62/234 (26%), Positives = 116/234 (49%), Gaps = 6/234 (2%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
E N+K ++S + E D+ N L+ K KLE L ++ +LE + R E+A
Sbjct: 998 EANKKTSDSLQAE-EDKCNHLN-------KLKAKLEQALDELEDNLER-EKKVRGDVEKA 1048
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLD------ESENNAMXXXX 367
K+ V +++ QE+ + ++++K++LE + +A++ E E N +
Sbjct: 1049 KRKVEQ------DLKSTQENVEDLERVKRELEENVRRKEAEISSLNSKLEDEQNLVSQLQ 1102
Query: 368 XXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVE 547
+ R+ ELE EL+AE+ + +K ++ R L+E+G + DE +L +
Sbjct: 1103 RKIKELQARIEELEEELEAERNARAKVEKQRAELNRELEELGERLDEAGGATSAQIELNK 1162
Query: 548 KLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTK 709
K + ++ +R +EEA ++ RK Q+ + E ADQ +Q LQK+++K
Sbjct: 1163 KREAELLKIRRDLEEASLQHEAQISALRKKHQDA--ANEMADQVDQ-LQKVKSK 1213
Score = 74.7 bits (182), Expect = 4e-13
Identities = 52/238 (21%), Positives = 110/238 (46%)
Frame = +2
Query: 5 RGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEA 184
R + E+ R E EL +RL+E +S+ + +K EA+L ++ DLEEAS +
Sbjct: 1123 RNARAKVEKQRAELNRELEELGERLDEAGGATSAQIELNKKREAELLKIRRDLEEASLQH 1182
Query: 185 RQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXX 364
+K D++ + + Q+ K+K +LE K+L+ ++D
Sbjct: 1183 EAQISALRKKHQDAA------NEMADQVDQLQKVKSKLEKDKKDLKREMD---------- 1226
Query: 365 XXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLV 544
+LES++ K ++K ++ E ++ ++ + ++ +++ LQ
Sbjct: 1227 ------------DLESQMTHNMKNKGCSEKVMKQFESQMSDLNARLEDSQRSINELQSQK 1274
Query: 545 EKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRS 718
+LQ + RQ+E+AE +V + ++ ++ED+ ++ +A KL+ + R+
Sbjct: 1275 SRLQAENSDLTRQLEDAEHRVSVLSKEKSQLSSQLEDARRSLEEETRARSKLQNEVRN 1332
Score = 70.5 bits (171), Expect = 7e-12
Identities = 52/251 (20%), Positives = 115/251 (45%), Gaps = 16/251 (6%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATK-------RKLEADLAAMQSDLEEA 172
++ ++ + E +K +L +++L Q + M K ++ E+ ++ + + LE++
Sbjct: 1204 VDQLQKVKSKLEKDKKDLKREMDDLESQMTHNMKNKGCSEKVMKQFESQMSDLNARLEDS 1263
Query: 173 SNEARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNA 352
+ Q + A++S L ++ + + K K QL +Q ++ + L+E
Sbjct: 1264 QRSINELQSQKSRLQAENSDLTRQLEDAEHRVSVLSKEKSQLSSQLEDARRSLEEETR-- 1321
Query: 353 MXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTD--------E 508
+ VR + +++DA +++ E Q++ ++R+L + + E
Sbjct: 1322 ------ARSKLQNEVRNMHADMDAIREQLEEEQESKSDVQRQLSKANNEIQQWRSKFESE 1375
Query: 509 DKKNQERLQDLVEKLQGKIKTYKRQVEEAE-EIAAVNLAKYRKIQQEIEDSEERADQAEQ 685
E L+D KL GK+ ++ E A + +A+ AK R +QQE+ED D+A
Sbjct: 1376 GANRTEELEDQKRKLLGKLSEAEQTTEAANAKCSALEKAKSR-LQQELEDMSIEVDRANA 1434
Query: 686 ALQKLRTKNRS 718
++ ++ K R+
Sbjct: 1435 SVNQMEKKQRA 1445
Score = 68.6 bits (166), Expect = 3e-11
Identities = 57/263 (21%), Positives = 115/263 (43%), Gaps = 37/263 (14%)
Frame = +2
Query: 11 SLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQ 190
S+ E+ ++ + E ++N L + + R A+L +++ +EE +
Sbjct: 1435 SVNQMEKKQRAFDKTTAEWQAKVNSLQSELENSQKESRGYSAELYRIKASIEEYQDSIGA 1494
Query: 191 ANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESEN-------- 346
+ K + L D++ + ++DK +++LE + +ELQA L+E+E
Sbjct: 1495 LRRENKNLADEIHDLTDQLSEGGRSTHELDKARRRLEMEKEELQAALEEAEGALEQEEAK 1554
Query: 347 --NAMXXXXXXXXXXEQRVRELESE------------------LDAEQKRHVETQKNTRK 466
A ++R++E E E L+AE K + + +K
Sbjct: 1555 VMRAQLEIATVRNEIDKRIQEKEEEFDNTRRNHQRALESMQASLEAEAKGKADAMRIKKK 1614
Query: 467 IERRLKEIGLQTD-------EDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAK 625
+E+ + E+ + D E +K +R Q + ++Q I+ +RQ +EA E + N+A+
Sbjct: 1615 LEQDINELEVALDASNRGKAEMEKTVKRYQQQIREMQTSIEEEQRQRDEARE--SYNMAE 1672
Query: 626 YR--KIQQEIEDSEERADQAEQA 688
R + E+E+ +QAE+A
Sbjct: 1673 RRCTLMSGEVEELRAALEQAERA 1695
Score = 60.5 bits (145), Expect = 7e-09
Identities = 54/264 (20%), Positives = 112/264 (42%), Gaps = 29/264 (10%)
Frame = +2
Query: 29 RNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQAK 208
R KN E +LTD+L+E + +R+LE + +Q+ LEEA Q +
Sbjct: 1497 RENKNLADEIHDLTDQLSEGGRSTHELDKARRRLEMEKEELQAALEEAEGALEQEEAKVM 1556
Query: 209 KAVADSSRLFDEI-RQEQEHAQQID----------------------------KIKKQLE 301
+A + + + +EI ++ QE ++ D +IKK+LE
Sbjct: 1557 RAQLEIATVRNEIDKRIQEKEEEFDNTRRNHQRALESMQASLEAEAKGKADAMRIKKKLE 1616
Query: 302 AQNKELQAKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRL 481
EL+ LD S N +Q++RE+++ ++ EQ++ E +++ ERR
Sbjct: 1617 QDINELEVALDAS-NRGKAEMEKTVKRYQQQIREMQTSIEEEQRQRDEARESYNMAERRC 1675
Query: 482 KEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSE 661
+ + +E + E+ + + ++ +V E + + RK++ +I +
Sbjct: 1676 TLMSGEVEELRAALEQAERARKASDNELADANDRVNELTSQVSSVQGQKRKLEGDINAMQ 1735
Query: 662 ERADQAEQALQKLRTKNRSSVSTA 733
D+ L+ + + +++ A
Sbjct: 1736 TDLDEMHGELKGADERCKKAMADA 1759
Score = 53.1 bits (126), Expect = 1e-06
Identities = 39/228 (17%), Positives = 98/228 (42%), Gaps = 10/228 (4%)
Frame = +2
Query: 11 SLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQ 190
S+ + + ++E +LT +L + + S K +L + L + LEE + +
Sbjct: 1266 SINELQSQKSRLQAENSDLTRQLEDAEHRVSVLSKEKSQLSSQLEDARRSLEEETRARSK 1325
Query: 191 ANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQ---AKLDESENNAMXX 361
+ + AD + +++ +EQE +++QL N E+Q +K + N
Sbjct: 1326 LQNEVRNMHADMDAIREQLEEEQESKSD---VQRQLSKANNEIQQWRSKFESEGANRTEE 1382
Query: 362 XXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTD-------EDKKN 520
++ E E +A + +K ++++ L+++ ++ D + +K
Sbjct: 1383 LEDQKRKLLGKLSEAEQTTEAANAKCSALEKAKSRLQQELEDMSIEVDRANASVNQMEKK 1442
Query: 521 QERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEE 664
Q + Q K+ + + ++E +++ + A+ +I+ IE+ ++
Sbjct: 1443 QRAFDKTTAEWQAKVNSLQSELENSQKESRGYSAELYRIKASIEEYQD 1490
>sp|P05661|MYSA_DROME Myosin heavy chain, muscle Length = 1962 Score = 278 bits (712), Expect = 1e-74 Identities = 142/250 (56%), Positives = 187/250 (74%) Frame = +2 Query: 5 RGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEA 184 R LE ++R R+ E E + ++LNE+S Q++S A KRKLE++L + SDL+E NEA Sbjct: 1688 RTLLEQADRGRRQAEQELADAHEQLNEVSAQNASISAAKRKLESELQTLHSDLDELLNEA 1747 Query: 185 RQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXX 364 + + E+AKKA+ D++RL DE+R EQ+HAQ +K++K LE Q KELQ +LDE+E NA+ Sbjct: 1748 KNSEEKAKKAMVDAARLADELRAEQDHAQTQEKLRKALEQQIKELQVRLDEAEANALKGG 1807 Query: 365 XXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLV 544 EQRVRELE+ELD EQ+RH + QKN RK ERR+KE+ Q++ED+KN ER+QDLV Sbjct: 1808 KKAIQKLEQRVRELENELDGEQRRHADAQKNLRKSERRVKELSFQSEEDRKNHERMQDLV 1867 Query: 545 EKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSV 724 +KLQ KIKTYKRQ+EEAEEIAA+NLAK+RK QQE+E++EERAD AEQA+ K R K R+ Sbjct: 1868 DKLQQKIKTYKRQIEEAEEIAALNLAKFRKAQQELEEAEERADLAEQAISKFRAKGRAG- 1926 Query: 725 STARGGSMAP 754 S RG S AP Sbjct: 1927 SVGRGASPAP 1936
Score = 77.4 bits (189), Expect = 5e-14
Identities = 53/222 (23%), Positives = 106/222 (47%), Gaps = 1/222 (0%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
LE R KN E +L D++ E +++LEA+ +Q+ LEEA Q
Sbjct: 1494 LEAVRRENKNLADEVKDLLDQIGEGGRNIHEIEKARKRLEAEKDELQAALEEAEAALEQE 1553
Query: 194 NEQAKKAVADSSRLFDEI-RQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXX 370
+ +A + S++ EI R+ QE ++ + +K + +QA L E+E
Sbjct: 1554 ENKVLRAQLELSQVRQEIDRRIQEKEEEFENTRKNHQRALDSMQASL-EAEAKGKAEALR 1612
Query: 371 XXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEK 550
E + ELE LD K + E QKN ++ +++LK+I +E+++ ++ ++ +
Sbjct: 1613 MKKKLEADINELEIALDHANKANAEAQKNIKRYQQQLKDIQTALEEEQRARDDAREQLGI 1672
Query: 551 LQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQ 676
+ + + ++EE+ + R+ +QE+ D+ E+ ++
Sbjct: 1673 SERRANALQNELEESRTLLEQADRGRRQAEQELADAHEQLNE 1714
Score = 75.1 bits (183), Expect = 3e-13
Identities = 51/238 (21%), Positives = 115/238 (48%), Gaps = 7/238 (2%)
Frame = +2
Query: 5 RGSLEN-SERNRKNT------ESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDL 163
+G+L++ ERN K T E++ ++ +RL + + K+K + +++ ++ D+
Sbjct: 894 KGALQDYQERNAKLTAQKNDLENQLRDIQERLTQEEDARNQLFQQKKKADQEISGLKKDI 953
Query: 164 EEASNEARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESE 343
E+ ++A + L DEI + E +++K KK N++ +L +E
Sbjct: 954 EDLELNVQKAEQDKATKDHQIRNLNDEIAHQDELINKLNKEKKMQGETNQKTGEELQAAE 1013
Query: 344 NNAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQ 523
+ + EQ + ELE L+ E+K + +K+ RK+E LK + ++N+
Sbjct: 1014 DK-INHLNKVKAKLEQTLDELEDSLEREKKVRGDVEKSKRKVEGDLKLTQEAVADLERNK 1072
Query: 524 ERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQK 697
+ L+ +++ ++ + ++E+ + + + + +++Q IE+ EE + QA K
Sbjct: 1073 KELEQTIQRKDKELSSITAKLEDEQVVVLKHQRQIKELQARIEELEEEVEAERQARAK 1130
Score = 67.8 bits (164), Expect = 4e-11
Identities = 54/256 (21%), Positives = 111/256 (43%), Gaps = 35/256 (13%)
Frame = +2
Query: 23 SERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQ 202
+E+ +K + E ++++L+ + + R +L ++ EE + +
Sbjct: 1441 AEKKQKAFDKIIGEWKLKVDDLAAELDASQKECRNYSTELFRLKGAYEEGQEQLEAVRRE 1500
Query: 203 AKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESEN----------NA 352
K + L D+I + + +I+K +K+LEA+ ELQA L+E+E A
Sbjct: 1501 NKNLADEVKDLLDQIGEGGRNIHEIEKARKRLEAEKDELQAALEEAEAALEQEENKVLRA 1560
Query: 353 MXXXXXXXXXXEQRVRELESE------------------LDAEQKRHVETQKNTRKIERR 478
++R++E E E L+AE K E + +K+E
Sbjct: 1561 QLELSQVRQEIDRRIQEKEEEFENTRKNHQRALDSMQASLEAEAKGKAEALRMKKKLEAD 1620
Query: 479 LKEIGLQTD-------EDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKI 637
+ E+ + D E +KN +R Q ++ +Q ++ +R ++A E ++ + +
Sbjct: 1621 INELEIALDHANKANAEAQKNIKRYQQQLKDIQTALEEEQRARDDAREQLGISERRANAL 1680
Query: 638 QQEIEDSEERADQAEQ 685
Q E+E+S +QA++
Sbjct: 1681 QNELEESRTLLEQADR 1696
Score = 66.6 bits (161), Expect = 1e-10
Identities = 64/253 (25%), Positives = 121/253 (47%), Gaps = 18/253 (7%)
Frame = +2
Query: 5 RGSLENSERNRKNTESEKVELTDRLN-ELSVQSSSFMATKRKLEA------DLAAMQSDL 163
+ + ++ RN + + + EL ++LN E +Q + T +L+A L +++ L
Sbjct: 967 KATKDHQIRNLNDEIAHQDELINKLNKEKKMQGETNQKTGEELQAAEDKINHLNKVKAKL 1026
Query: 164 EEASNEARQANEQAKKAVAD----SSRLFDEIRQEQEHAQQIDKIKKQLEA----QNKEL 319
E+ +E + E+ KK D ++ +++ QE +++ KK+LE ++KEL
Sbjct: 1027 EQTLDELEDSLEREKKVRGDVEKSKRKVEGDLKLTQEAVADLERNKKELEQTIQRKDKEL 1086
Query: 320 Q---AKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEI 490
AKL E E + + R+ ELE E++AE++ + +K + R L+E+
Sbjct: 1087 SSITAKL-EDEQVVVLKHQRQIKELQARIEELEEEVEAERQARAKAEKQRADLARELEEL 1145
Query: 491 GLQTDEDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERA 670
G + +E +L +K + ++ +R +EEA LA RK + + E A
Sbjct: 1146 GERLEEAGGATSAQIELNKKREAELSKLRRDLEEANIQHESTLANLRK--KHNDAVAEMA 1203
Query: 671 DQAEQALQKLRTK 709
+Q +Q L KL+ K
Sbjct: 1204 EQVDQ-LNKLKAK 1215
Score = 46.6 bits (109), Expect = 1e-04
Identities = 57/247 (23%), Positives = 104/247 (42%), Gaps = 16/247 (6%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANE-- 199
E+ K + E+ +L+E + + F A+K+KL + + + LEEA ++ Q ++
Sbjct: 1245 EKIAKQLQHTLNEVQSKLDETNRTLNDFDASKKKLSIENSDLLRQLEEAESQVSQLSKIK 1304
Query: 200 -QAKKAVADSSRLFDEIRQEQ----------EHAQQIDKIKKQLEAQ---NKELQAKLDE 337
+ D+ RL DE +E+ EH +D +++Q+E + +LQ +L +
Sbjct: 1305 ISLTTQLEDTKRLADEESRERATLLGKFRNLEH--DLDNLREQVEEEAEGKADLQRQLSK 1362
Query: 338 SENNAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKK 517
+ A R ELE Q R E ++ + + K IGL+ K
Sbjct: 1363 ANAEAQVWRSKYESDGVARSEELEEAKRKLQARLAEAEETIESLNQ--KCIGLE-----K 1415
Query: 518 NQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQK 697
++RL VE LQ +V+ A IA K + + I + + + D L
Sbjct: 1416 TKQRLSTEVEDLQ-------LEVDRANAIANAAEKKQKAFDKIIGEWKLKVDDLAAELDA 1468
Query: 698 LRTKNRS 718
+ + R+
Sbjct: 1469 SQKECRN 1475
Score = 43.5 bits (101), Expect = 9e-04
Identities = 45/167 (26%), Positives = 78/167 (46%), Gaps = 12/167 (7%)
Frame = +2
Query: 200 QAKKAVADSSRLFDEIRQEQEHAQQID-------KIKKQLEAQNKELQAKLDESENNAMX 358
Q K + + SR+ DEI + +E A++ + K++K+LEA N +L A E A+
Sbjct: 833 QKVKPLLNVSRIEDEIARLEEKAKKAEELHAAEVKVRKELEALNAKLLA-----EKTALL 887
Query: 359 XXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQ----- 523
Q +E ++L A QK +E Q R I+ RL + +ED +NQ
Sbjct: 888 DSLSGEKGALQDYQERNAKLTA-QKNDLENQ--LRDIQERLTQ-----EEDARNQLFQQK 939
Query: 524 ERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEE 664
++ + L+ I+ + V++AE+ A + R + EI +E
Sbjct: 940 KKADQEISGLKKDIEDLELNVQKAEQDKATKDHQIRNLNDEIAHQDE 986
>sp|P02566|MYO4_CAEEL Myosin-4 (Myosin heavy chain B) (MHC B) (Uncoordinated protein 54) Length = 1966 Score = 251 bits (640), Expect = 3e-66 Identities = 125/241 (51%), Positives = 176/241 (73%) Frame = +2 Query: 17 ENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQAN 196 E +ER RK E E + D+ NE + Q SS + KRKLE ++ A+ +DL+E NE + A Sbjct: 1705 EAAERARKQAEYEAADARDQANEANAQVSSLTSAKRKLEGEIQAIHADLDETLNEYKAAE 1764 Query: 197 EQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXX 376 E++KKA+AD++RL +E+RQEQEH+Q +D+++K LE Q KE+Q +LDE+E A+ Sbjct: 1765 ERSKKAIADATRLAEELRQEQEHSQHVDRLRKGLEQQLKEIQVRLDEAEAAALKGGKKVI 1824 Query: 377 XXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQ 556 EQRVRELESELD EQ+R + KN + +RR++E+ Q DEDKKN ERLQDL++KLQ Sbjct: 1825 AKLEQRVRELESELDGEQRRFQDANKNLGRADRRVRELQFQVDEDKKNFERLQDLIDKLQ 1884 Query: 557 GKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVSTAR 736 K+KT K+QVEEAEE+A +NL KY+++ ++ED+EERADQAE +L K+R+K+R+S S A Sbjct: 1885 QKLKTQKKQVEEAEELANLNLQKYKQLTHQLEDAEERADQAENSLSKMRSKSRASASVAP 1944 Query: 737 G 739 G Sbjct: 1945 G 1945
Score = 72.8 bits (177), Expect = 1e-12
Identities = 57/226 (25%), Positives = 99/226 (43%), Gaps = 1/226 (0%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
+E R K+ E +LTD+L E R+LE + +Q L+EA
Sbjct: 1507 VEGLRRENKSLSQEIKDLTDQLGEGGRSVHEMQKIIRRLEIEKEELQHALDEAEAALEAE 1566
Query: 194 NEQAKKAVADSSRLFDEIRQE-QEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXX 370
+ +A + S++ EI + QE ++ + +K + +QA L E+E
Sbjct: 1567 ESKVLRAQVEVSQIRSEIEKRIQEKEEEFENTRKNHARALESMQASL-ETEAKGKAELLR 1625
Query: 371 XXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEK 550
E + ELE LD K + + QKN ++ + +++E+ LQ +E+++N D E+
Sbjct: 1626 IKKKLEGDINELEIALDHANKANADAQKNLKRYQEQVRELQLQVEEEQRNG---ADTREQ 1682
Query: 551 LQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQA 688
K E EE+ N A R +Q ++ + DQA +A
Sbjct: 1683 FFNAEKRATLLQSEKEELLVANEAAERARKQAEYEAADARDQANEA 1728
Score = 60.5 bits (145), Expect = 7e-09
Identities = 56/226 (24%), Positives = 101/226 (44%), Gaps = 9/226 (3%)
Frame = +2
Query: 35 RKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQAKKA 214
RK T+ EL +L S+ K Q +L E R+ N+ +
Sbjct: 1469 RKKTDDLAAELDGAQRDLRNTSTDLFKAKNA--------QEELAEVVEGLRRENKSLSQE 1520
Query: 215 VADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXXEQR 394
+ D L D++ + ++ KI ++LE + +ELQ LDE+E A+ +
Sbjct: 1521 IKD---LTDQLGEGGRSVHEMQKIIRRLEIEKEELQHALDEAE-AALEAEESKVLRAQVE 1576
Query: 395 VRELESELDAEQKRHVETQKNTRKIE-RRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKT 571
V ++ SE++ + E +NTRK R L+ + + + K + L + +KL+G I
Sbjct: 1577 VSQIRSEIEKRIQEKEEEFENTRKNHARALESMQASLETEAKGKAELLRIKKKLEGDINE 1636
Query: 572 YKRQVEEAEEI---AAVNLAKY----RKIQQEIEDSEER-ADQAEQ 685
+ ++ A + A NL +Y R++Q ++E+ + AD EQ
Sbjct: 1637 LEIALDHANKANADAQKNLKRYQEQVRELQLQVEEEQRNGADTREQ 1682
Score = 58.2 bits (139), Expect = 3e-08
Identities = 53/269 (19%), Positives = 106/269 (39%), Gaps = 28/269 (10%)
Frame = +2
Query: 11 SLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQ 190
SL +E +++ + + L D + + + K+ E + DL+ ++
Sbjct: 972 SLRKAESEKQSKDHQIRSLQDEMQQQDEAIAKLNKEKKHQEEINRKLMEDLQSEEDKGNH 1031
Query: 191 ANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDES-------ENN 349
N+ K L D + +E+ +DK K+++E + K Q +DES ENN
Sbjct: 1032 QNKVKAKLEQTLDDLEDSLEREKRARADLDKQKRKVEGELKIAQENIDESGRQRHDLENN 1091
Query: 350 --------------------AMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKI 469
+ + R+ ELE EL+ E++ + + +
Sbjct: 1092 LKKKESELHSVSSRLEDEQALVSKLQRQIKDGQSRISELEEELENERQSRSKADRAKSDL 1151
Query: 470 ERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQE- 646
+R L+E+G + DE ++ +K + ++ +R +EEA L RK +
Sbjct: 1152 QRELEELGEKLDEQGGATAAQVEVNKKREAELAKLRRDLEEANMNHENQLGGLRKKHTDA 1211
Query: 647 IEDSEERADQAEQALQKLRTKNRSSVSTA 733
+ + ++ DQ +A K+ +V A
Sbjct: 1212 VAELTDQLDQLNKAKAKVEKDKAQAVRDA 1240
Score = 32.3 bits (72), Expect = 2.0
Identities = 20/71 (28%), Positives = 37/71 (52%), Gaps = 9/71 (12%)
Frame = +2
Query: 548 KLQGKIKTYKRQVEEAEEIAAVN---------LAKYRKIQQEIEDSEERADQAEQALQKL 700
KL GK+K + +EAEE+ +N LAK K+++E+E+S + + + +L
Sbjct: 843 KLYGKVKPMLKAGKEAEELEKINDKVKALEDSLAKEEKLRKELEESSAKLVEEKTSLFTN 902
Query: 701 RTKNRSSVSTA 733
++ +S A
Sbjct: 903 LESTKTQLSDA 913
>sp|P12844|MYO3_CAEEL Myosin-3 (Myosin heavy chain A) (MHC A) Length = 1969 Score = 236 bits (602), Expect = 7e-62 Identities = 121/239 (50%), Positives = 170/239 (71%) Frame = +2 Query: 17 ENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQAN 196 E +ER R+N E+E +EL ++ N+L+ S+ +RKLE +L A ++LEE +NE + A Sbjct: 1706 EAAERARRNAEAECIELREQNNDLNAHVSALTGQRRKLEGELLAAHAELEEIANELKNAV 1765 Query: 197 EQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXX 376 EQ +KA AD++RL +E+RQEQEH+ I++I+K LE Q KE+Q +LD++EN A+ Sbjct: 1766 EQGQKASADAARLAEELRQEQEHSMHIERIRKGLELQIKEMQIRLDDAENAALKGGKKII 1825 Query: 377 XXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQ 556 E R+R +E ELD EQ+RH +T+KN RK ERR+KE+ Q E+KKN+ERL +LV+KLQ Sbjct: 1826 AQLEARIRAIEQELDGEQRRHQDTEKNWRKAERRVKEVEFQVVEEKKNEERLTELVDKLQ 1885 Query: 557 GKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVSTA 733 K+K +KRQVEEAEE+AA NL KY+ + + E +EERAD AE AL K+R K R+S S A Sbjct: 1886 CKLKIFKRQVEEAEEVAASNLNKYKVLTAQFEQAEERADIAENALSKMRNKIRASASMA 1944
Score = 70.9 bits (172), Expect = 5e-12
Identities = 53/238 (22%), Positives = 110/238 (46%), Gaps = 6/238 (2%)
Frame = +2
Query: 26 ERNRK------NTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEAR 187
ERN K ES+ ++T +L ++ ++ K+K + +L+ + +++ R
Sbjct: 916 ERNEKLNQLKATLESKLSDITGQLEDMQERNEDLARQKKKTDQELSDTKKHVQDLELSLR 975
Query: 188 QANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXX 367
+A ++ + + L DE+ + E +++K KK E N++L L +SE + +
Sbjct: 976 KAEQEKQSRDHNIRSLQDEMANQDEAVAKLNKEKKHQEESNRKLNEDL-QSEEDKVNHLE 1034
Query: 368 XXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVE 547
EQ++ ELE +D E++ + +K RK+E LK DE K + ++ ++
Sbjct: 1035 KIRNKLEQQMDELEENIDREKRSRGDIEKAKRKVEGDLKVAQENIDEITKQKHDVETTLK 1094
Query: 548 KLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSS 721
+ + + ++ E I A K+Q+ I++ R + E+ L+ R + S
Sbjct: 1095 RKEEDLHHTNAKLAENNSIIA-------KLQRLIKELTARNAELEEELEAERNSRQKS 1145
Score = 65.9 bits (159), Expect = 2e-10
Identities = 55/236 (23%), Positives = 109/236 (46%), Gaps = 3/236 (1%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
L+++ R K+ E +LTD+L E + RKLE + +Q L+EA EA
Sbjct: 1508 LDSTRRENKSLAQEVKDLTDQLGEGGRSVAELQKIVRKLEVEKEELQKALDEA--EAALE 1565
Query: 194 NEQAK--KAVADSSRLFDEIRQE-QEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXX 364
E+AK +A + S++ EI + QE ++ + ++ + + +QA L E+E
Sbjct: 1566 AEEAKVLRAQIEVSQIRSEIEKRIQEKEEEFENTRRNHQRALESMQATL-EAETKQKEEA 1624
Query: 365 XXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLV 544
E + +LE LD + + + QK +K ++E+ Q +E+++ ++ +++
Sbjct: 1625 LRIKKKLESDINDLEIALDHANRAYADAQKTIKKYMETVQELQFQIEEEQRQKDEIRE-- 1682
Query: 545 EKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKN 712
Q +E+ A+ ++ ++ Q+ E +E AE +LR +N
Sbjct: 1683 ------------QFLASEKRNAILQSEKDELAQQAEAAERARRNAEAECIELREQN 1726
Score = 65.1 bits (157), Expect = 3e-10
Identities = 55/256 (21%), Positives = 112/256 (43%), Gaps = 35/256 (13%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
E++R+ ES E + ++LS + + R+L DL ++ +E + +
Sbjct: 1456 EKHRRQFESIIAEWKKKTDDLSSELDAAQRDNRQLSTDLFKAKTANDELAEYLDSTRREN 1515
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESEN----------NAM 355
K + L D++ + ++ KI ++LE + +ELQ LDE+E A
Sbjct: 1516 KSLAQEVKDLTDQLGEGGRSVAELQKIVRKLEVEKEELQKALDEAEAALEAEEAKVLRAQ 1575
Query: 356 XXXXXXXXXXEQRVRELESE------------------LDAEQKRHVETQKNTRKIERRL 481
E+R++E E E L+AE K+ E + +K+E +
Sbjct: 1576 IEVSQIRSEIEKRIQEKEEEFENTRRNHQRALESMQATLEAETKQKEEALRIKKKLESDI 1635
Query: 482 KEIGLQTD-------EDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQ 640
++ + D + +K ++ + V++LQ +I+ +RQ +E E + + +Q
Sbjct: 1636 NDLEIALDHANRAYADAQKTIKKYMETVQELQFQIEEEQRQKDEIREQFLASEKRNAILQ 1695
Query: 641 QEIEDSEERADQAEQA 688
E ++ ++A+ AE+A
Sbjct: 1696 SEKDELAQQAEAAERA 1711
Score = 54.3 bits (129), Expect = 5e-07
Identities = 62/252 (24%), Positives = 109/252 (43%), Gaps = 14/252 (5%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEA--- 184
LE E +K + + ELTD L + +S + KL DL QSD+E+A+ +
Sbjct: 1396 LEEIEAAKKALQLKVQELTDTNEGLFAKIASQEKVRFKLMQDLDDAQSDVEKAAAQVAFY 1455
Query: 185 RQANEQAKKAVADSSRLFDEIRQEQEHAQ----QIDKIKKQLEAQNKELQAKLDES--EN 346
+ Q + +A+ + D++ E + AQ Q+ + + N EL LD + EN
Sbjct: 1456 EKHRRQFESIIAEWKKKTDDLSSELDAAQRDNRQLSTDLFKAKTANDELAEYLDSTRREN 1515
Query: 347 NAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDE-----D 511
++ Q V++L +L + E QK RK+E +E+ DE +
Sbjct: 1516 KSL----------AQEVKDLTDQLGEGGRSVAELQKIVRKLEVEKEELQKALDEAEAALE 1565
Query: 512 KKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQAL 691
+ + L+ +E Q + + KR E+ EE R ++ E Q E+AL
Sbjct: 1566 AEEAKVLRAQIEVSQIRSEIEKRIQEKEEEFENTRRNHQRALESMQATLEAETKQKEEAL 1625
Query: 692 QKLRTKNRSSVS 727
+++ K S ++
Sbjct: 1626 -RIKKKLESDIN 1636
Score = 51.6 bits (122), Expect = 3e-06
Identities = 61/237 (25%), Positives = 107/237 (45%), Gaps = 10/237 (4%)
Frame = +2
Query: 17 ENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQAN 196
E E R+N + + L + Q + K+KLE+D+ +DLE A + A +A
Sbjct: 1594 EEFENTRRNHQRALESMQATLEAETKQKEEALRIKKKLESDI----NDLEIALDHANRAY 1649
Query: 197 EQAKKAVADSSRLFDEIR-QEQEHAQQIDKIKKQL---EAQNKELQAKLDESENNAMXXX 364
A+K + E++ Q +E +Q D+I++Q E +N LQ++ DE A
Sbjct: 1650 ADAQKTIKKYMETVQELQFQIEEEQRQKDEIREQFLASEKRNAILQSEKDELAQQAEAAE 1709
Query: 365 XXXXXXXEQ--RVRELESELDAEQKRHVET-QKNTRKIERRLKEIGLQTDEDKKNQERLQ 535
+ +RE ++L+A HV RK+E L + +E L+
Sbjct: 1710 RARRNAEAECIELREQNNDLNA----HVSALTGQRRKLEGELLAAHAELEE---IANELK 1762
Query: 536 DLVEKLQGKIKTYKRQVEE--AEEIAAVNLAKYRK-IQQEIEDSEERADQAEQALQK 697
+ VE+ Q R EE E+ ++++ + RK ++ +I++ + R D AE A K
Sbjct: 1763 NAVEQGQKASADAARLAEELRQEQEHSMHIERIRKGLELQIKEMQIRLDDAENAALK 1819
>sp|P02565|MYH3_CHICK Myosin heavy chain, fast skeletal muscle, embryonic Length = 1940 Score = 232 bits (591), Expect = 1e-60 Identities = 116/238 (48%), Positives = 174/238 (73%) Frame = +2 Query: 2 VRGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNE 181 +R +LE +ER+RK E E ++ ++R+ L Q++S + TK+KLE+D++ +QS++E+ E Sbjct: 1693 LRAALEQTERSRKVAEQELLDASERVQLLHTQNTSLINTKKKLESDISQIQSEMEDTIQE 1752 Query: 182 ARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXX 361 AR A E+AKKA+ D++ + +E+++EQ+ + ++++KK L+ K+LQ +LDE+E A+ Sbjct: 1753 ARNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNLDQTVKDLQHRLDEAEQLALKG 1812 Query: 362 XXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDL 541 E RVRELE E+DAEQKR E K RK ERR+KE+ Q++ED+KN RLQDL Sbjct: 1813 GKKQIQKLEARVRELEGEVDAEQKRSAEAVKGVRKYERRVKELTYQSEEDRKNVLRLQDL 1872 Query: 542 VEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNR 715 V+KLQ K+K+YKRQ EEAEE++ VNL+K+RKIQ E+E++EERAD AE + KLR K+R Sbjct: 1873 VDKLQMKVKSYKRQAEEAEELSNVNLSKFRKIQHELEEAEERADIAESQVNKLRAKSR 1930
Score = 72.4 bits (176), Expect = 2e-12
Identities = 50/227 (22%), Positives = 113/227 (49%)
Frame = +2
Query: 47 ESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQAKKAVADS 226
E++ ELT+R + ++ A KRKLE + + ++ D+++ + ++
Sbjct: 921 EAKIKELTERAEDEEEMNAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKV 980
Query: 227 SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXXEQRVREL 406
L +E+ E ++ K KK L+ +++ L ++E + + EQ+V +L
Sbjct: 981 KNLTEEMAALDETIAKLTKEKKALQEAHQQTLDDL-QAEEDKVNTLTKAKTKLEQQVDDL 1039
Query: 407 ESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQV 586
E L+ E+K ++ ++ RK+E LK T + + ++++L + ++K +I + ++
Sbjct: 1040 EGSLEQEKKLRMDLERAKRKLEGDLKMTQESTMDLENDKQQLDEKLKKKDFEISQIQSKI 1099
Query: 587 EEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVS 727
E+ + + K +++Q IE+ EE +AE+ + K+R+ +S
Sbjct: 1100 EDEQALGMQLQKKIKELQARIEELEEEI-EAERTSRAKAEKHRADLS 1145
Score = 70.5 bits (171), Expect = 7e-12
Identities = 58/254 (22%), Positives = 112/254 (44%), Gaps = 10/254 (3%)
Frame = +2
Query: 5 RGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEA 184
R S +E++R + E E+++RL E +++ + +K EA+ M+ DLEEA+ +
Sbjct: 1131 RTSRAKAEKHRADLSRELEEISERLEEAGGATAAQIDMNKKREAEFQKMRRDLEEATLQH 1190
Query: 185 RQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXX 364
+K ADS+ E + ++K++LE + EL+ ++D+ +N M
Sbjct: 1191 EATAAALRKKHADST------ADVGEQIDNLQRVKQKLEKEKSELKMEIDDLASN-MESV 1243
Query: 365 XXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLV 544
E+ R LE +L + + E Q+ I + + ++ E + E L+
Sbjct: 1244 SKAKANLEKMCRSLEDQLSEIKTKEEEQQRTINDISAQKARLQTESGEYSRQVEEKDALI 1303
Query: 545 EKLQGKIKTYKRQVEE----------AEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQ 694
+L + + +Q+EE A++ A L R + + E +A+ LQ
Sbjct: 1304 SQLSRGKQAFTQQIEELKRHLEEEIKAKKCPAHALQSARHDCDLLREQYEEEQEAKGELQ 1363
Query: 695 KLRTKNRSSVSTAR 736
+ +K S V+ R
Sbjct: 1364 RALSKANSEVAQWR 1377
Score = 67.4 bits (163), Expect = 6e-11
Identities = 53/235 (22%), Positives = 112/235 (47%), Gaps = 14/235 (5%)
Frame = +2
Query: 17 ENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQAN 196
E+ E ++K +L D +EL T K+E + A ++ ++ + E +
Sbjct: 932 EDEEEMNAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAALD 991
Query: 197 E------QAKKAVADS-SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAM 355
E + KKA+ ++ + D+++ E++ + K K +LE Q +L+ L++ + M
Sbjct: 992 ETIAKLTKEKKALQEAHQQTLDDLQAEEDKVNTLTKAKTKLEQQVDDLEGSLEQEKKLRM 1051
Query: 356 XXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQ 535
E+ R+LE +L Q+ ++ + + ++++ +LK+ + + + E Q
Sbjct: 1052 DL--------ERAKRKLEGDLKMTQESTMDLENDKQQLDEKLKKKDFEISQIQSKIEDEQ 1103
Query: 536 DLVEKLQGKIKTYKRQVE------EAEEIAAVNLAKYR-KIQQEIEDSEERADQA 679
L +LQ KIK + ++E EAE + K+R + +E+E+ ER ++A
Sbjct: 1104 ALGMQLQKKIKELQARIEELEEEIEAERTSRAKAEKHRADLSRELEEISERLEEA 1158
Score = 63.9 bits (154), Expect = 6e-10
Identities = 61/286 (21%), Positives = 111/286 (38%), Gaps = 49/286 (17%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
++ +KN + E + E + + R L +L M++ EE+ + +
Sbjct: 1448 DKKQKNFDKILSEWKQKYEETQAELEASQKESRSLSTELFKMKNAYEESLDHLETLKREN 1507
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESE-------------- 343
K + S L ++I + + +++K+KKQ+E + ELQ L+E+E
Sbjct: 1508 KNLQQEISDLTEQIAEGGKAIHELEKVKKQIEQEKSELQTALEEAEASLEHEEGKILRVQ 1567
Query: 344 ---NNAMXXXXXXXXXXEQRVREL-----------ESELDAEQKRHVETQKNTRKIERRL 481
N ++ + +L +S LDAE + E + +K+E L
Sbjct: 1568 LELNQVKSDIDRKIAEKDEEIDQLKRNHLRVVDSMQSTLDAEIRSRNEALRLKKKMEGDL 1627
Query: 482 KEIGLQTDEDKKNQERLQDLVEKLQGKIK--------------TYKRQVEEAEEIAAVNL 619
EI +Q + Q + QG +K K QV E A +
Sbjct: 1628 NEIEIQLSHANRQAAEAQKNLRNTQGVLKDTQIHLDDALRSQEDLKEQVAMVERRANLLQ 1687
Query: 620 AKYRKIQQEIEDSEERADQAEQAL-------QKLRTKNRSSVSTAR 736
A+ +++ +E +E AEQ L Q L T+N S ++T +
Sbjct: 1688 AEIEELRAALEQTERSRKVAEQELLDASERVQLLHTQNTSLINTKK 1733
Score = 52.0 bits (123), Expect = 2e-06
Identities = 57/230 (24%), Positives = 100/230 (43%), Gaps = 4/230 (1%)
Frame = +2
Query: 38 KNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQAKKAV 217
K+ ESEK E++ F TK +L A A + +LEE Q + V
Sbjct: 846 KSAESEK--------EMANMKEEFEKTKEEL-AKSEAKRKELEEKMVSLLQEKNDLQLQV 896
Query: 218 ADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKL-DESENNAMXXXXXXXXXXEQR 394
+ D + +E Q+ K K QLEA+ KEL + DE E NA E
Sbjct: 897 QAEA---DGLADAEERCDQLIKTKIQLEAKIKELTERAEDEEEMNA--ELTAKKRKLEDE 951
Query: 395 VRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTY 574
EL+ ++D + + +K E ++K + T+E E + L ++ + + +
Sbjct: 952 CSELKKDIDDLELTLAKVEKEKHATENKVKNL---TEEMAALDETIAKLTKEKKALQEAH 1008
Query: 575 KRQVE--EAEEIAAVNLAKYR-KIQQEIEDSEERADQAEQALQKLRTKNR 715
++ ++ +AEE L K + K++Q+++D E +Q ++ L R
Sbjct: 1009 QQTLDDLQAEEDKVNTLTKAKTKLEQQVDDLEGSLEQEKKLRMDLERAKR 1058
Score = 46.6 bits (109), Expect = 1e-04
Identities = 54/286 (18%), Positives = 112/286 (39%), Gaps = 49/286 (17%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
++N +R ++ E EK EL +++L+ S K LE +++ L E + +
Sbjct: 1212 IDNLQRVKQKLEKEKSELKMEIDDLASNMESVSKAKANLEKMCRSLEDQLSEIKTKEEEQ 1271
Query: 194 NEQAKKAVADSSRLFDEI------------------RQEQEHAQQIDKIKKQLEAQNK-- 313
A +RL E R +Q QQI+++K+ LE + K
Sbjct: 1272 QRTINDISAQKARLQTESGEYSRQVEEKDALISQLSRGKQAFTQQIEELKRHLEEEIKAK 1331
Query: 314 --------------ELQAKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQ-KRHVET 448
+L + E E A V + ++ + + +R E
Sbjct: 1332 KCPAHALQSARHDCDLLREQYEEEQEAKGELQRALSKANSEVAQWRTKYETDAIQRTEEL 1391
Query: 449 QKNTRKIERRLKE-------IGLQTDEDKKNQERLQDLVEKLQGKIK-------TYKRQV 586
++ +K+ +RL++ + + +K ++RLQ+ VE L ++ ++
Sbjct: 1392 EEAKKKLAQRLQDAEEHVEAVNSKCASLEKTKQRLQNEVEDLMIDVERSNAACAALDKKQ 1451
Query: 587 EEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSV 724
+ ++I + KY + Q E+E S++ + L K++ S+
Sbjct: 1452 KNFDKILSEWKQKYEETQAELEASQKESRSLSTELFKMKNAYEESL 1497
Score = 40.4 bits (93), Expect = 0.007
Identities = 32/233 (13%), Positives = 93/233 (39%)
Frame = +2
Query: 20 NSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANE 199
++++ R TES E + ++ E S K+ + ++ LEE +
Sbjct: 1279 SAQKARLQTESG--EYSRQVEEKDALISQLSRGKQAFTQQIEELKRHLEEEIKAKKCPAH 1336
Query: 200 QAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXX 379
+ A D L ++ +EQE ++ + + ++ + + K +
Sbjct: 1337 ALQSARHDCDLLREQYEEEQEAKGELQRALSKANSEVAQWRTKYETDAIQRTEELEEAKK 1396
Query: 380 XXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQG 559
QR+++ E ++A + +K ++++ ++++ + + L +
Sbjct: 1397 KLAQRLQDAEEHVEAVNSKCASLEKTKQRLQNEVEDLMIDVERSNAACAALDKKQKNFDK 1456
Query: 560 KIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRS 718
+ +K++ EE + + + R + E+ + +++ L+ L+ +N++
Sbjct: 1457 ILSEWKQKYEETQAELEASQKESRSLSTELFKMKNAYEESLDHLETLKRENKN 1509
Score = 37.7 bits (86), Expect = 0.048
Identities = 25/119 (21%), Positives = 55/119 (46%), Gaps = 7/119 (5%)
Frame = +2
Query: 386 EQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKN----QERLQDLVE-- 547
++ + + EL + + E ++ + + ++ LQ + +ER L++
Sbjct: 858 KEEFEKTKEELAKSEAKRKELEEKMVSLLQEKNDLQLQVQAEADGLADAEERCDQLIKTK 917
Query: 548 -KLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSS 721
+L+ KIK + E+ EE+ A AK RK++ E + ++ D E L K+ + ++
Sbjct: 918 IQLEAKIKELTERAEDEEEMNAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHAT 976
>sp|P13538|MYSS_CHICK Myosin heavy chain, skeletal muscle, adult Length = 1939 Score = 231 bits (588), Expect = 3e-60 Identities = 115/238 (48%), Positives = 171/238 (71%) Frame = +2 Query: 2 VRGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNE 181 +RG+LE +ER+RK E E ++ T+R+ L Q++S + TK+KLE D+ +QS++E+ E Sbjct: 1691 LRGALEQTERSRKVAEQELLDATERVQLLHTQNTSLINTKKKLETDIVQIQSEMEDTIQE 1750 Query: 182 ARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXX 361 AR A E+AKKA+ D++ + +E+++EQ+ + ++++KK ++ K+L +LDE+E A+ Sbjct: 1751 ARNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNMDQTVKDLHVRLDEAEQLALKG 1810 Query: 362 XXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDL 541 E RVRELE E+D+EQKR E K RK ERR+KE+ Q +ED+KN RLQDL Sbjct: 1811 GKKQLQKLEARVRELEGEVDSEQKRSAEAVKGVRKYERRVKELTYQCEEDRKNILRLQDL 1870 Query: 542 VEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNR 715 V+KLQ K+K+YKRQ EEAEE++ VNL+K+RKIQ E+E++EERAD AE + KLR K+R Sbjct: 1871 VDKLQMKVKSYKRQAEEAEELSNVNLSKFRKIQHELEEAEERADIAESQVNKLRVKSR 1928
Score = 73.9 bits (180), Expect = 6e-13
Identities = 57/242 (23%), Positives = 111/242 (45%), Gaps = 1/242 (0%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
LE +R KN + E +LT+++ E K+ +E + + +Q+ LEEA
Sbjct: 1498 LETLKRENKNLQQEIADLTEQIAEGGKAVHELEKVKKHVEQEKSELQASLEEAEASLEHE 1557
Query: 194 NEQAKKAVADSSRLFDEI-RQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXX 370
+ + + +++ EI R+ E ++ID++K+ + +Q+ LD +E +
Sbjct: 1558 EGKILRLQLELNQIKSEIDRKIAEKDEEIDQLKRNHLRIVESMQSTLD-AEIRSRNEALR 1616
Query: 371 XXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEK 550
E + E+E +L + E QKN R + LK+ + D+ + QE L++ V
Sbjct: 1617 LKKKMEGDLNEMEIQLSHANRMAAEAQKNLRNTQGTLKDTQIHLDDALRTQEDLKEQVAM 1676
Query: 551 LQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVST 730
++ + + +VEE + +QE+ D+ ER +Q L T+N S ++T
Sbjct: 1677 VERRANLLQAEVEELRGALEQTERSRKVAEQELLDATER-------VQLLHTQNTSLINT 1729
Query: 731 AR 736
+
Sbjct: 1730 KK 1731
Score = 72.8 bits (177), Expect = 1e-12
Identities = 60/254 (23%), Positives = 115/254 (45%), Gaps = 10/254 (3%)
Frame = +2
Query: 5 RGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEA 184
R S +E++R + E E+++RL E +++ + +K EA+ M+ DLEEA+ +
Sbjct: 1129 RTSRAKAEKHRADLSRELEEISERLEEAGGATAAQIEMNKKREAEFQKMRRDLEEATLQH 1188
Query: 185 RQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXX 364
+K ADS+ + E + ++K++LE + EL+ ++D+ +N M
Sbjct: 1189 EATAAALRKKHADST------AELGEQIDNLQRVKQKLEKEKSELKMEIDDLASN-MESV 1241
Query: 365 XXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLV 544
E+ R LE +L + + + Q+ + + + +T E + E L+
Sbjct: 1242 SKAKANLEKMCRTLEDQLSEIKTKEEQNQRMINDLNTQRARLQTETGEYSRQAEEKDALI 1301
Query: 545 EKLQGKIKTYKRQVEE-----AEEIAAVN-----LAKYRKIQQEIEDSEERADQAEQALQ 694
+L + + +Q+EE EEI A N L R + + + E +A+ LQ
Sbjct: 1302 SQLSRGKQGFTQQIEELKRHLEEEIKAKNALAHALQSARHDCELLREQYEEEQEAKGELQ 1361
Query: 695 KLRTKNRSSVSTAR 736
+ +K S V+ R
Sbjct: 1362 RALSKANSEVAQWR 1375
Score = 65.5 bits (158), Expect = 2e-10
Identities = 48/227 (21%), Positives = 111/227 (48%)
Frame = +2
Query: 47 ESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQAKKAVADS 226
E++ E+T+R + ++ A KRKLE + + ++ D+++ + ++
Sbjct: 919 EAKIKEVTERAEDEEEINAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKV 978
Query: 227 SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXXEQRVREL 406
L +E+ E ++ K KK L+ +++ L + E + + EQ+V +L
Sbjct: 979 KNLTEEMAVLDETIAKLTKEKKALQEAHQQTLDDL-QVEEDKVNTLTKAKTKLEQQVDDL 1037
Query: 407 ESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQV 586
E L+ E+K ++ ++ RK+E LK + + ++++L + ++K +I + ++
Sbjct: 1038 EGSLEQEKKLRMDLERAKRKLEGDLKLAHDSIMDLENDKQQLDEKLKKKDFEISQIQSKI 1097
Query: 587 EEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVS 727
E+ + + K +++Q IE+ EE +AE+ + K+R+ +S
Sbjct: 1098 EDEQALGMQLQKKIKELQARIEELEEEI-EAERTSRAKAEKHRADLS 1143
Score = 64.3 bits (155), Expect = 5e-10
Identities = 46/229 (20%), Positives = 105/229 (45%), Gaps = 3/229 (1%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
++ +KN + E + E + + R L +L M++ EE+ + +
Sbjct: 1446 DKKQKNFDKILAEWKQKYEETQTELEASQKESRSLSTELFKMKNAYEESLDHLETLKREN 1505
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXX 385
K + + L ++I + + +++K+KK +E + ELQA L+E+E ++
Sbjct: 1506 KNLQQEIADLTEQIAEGGKAVHELEKVKKHVEQEKSELQASLEEAE-ASLEHEEGKILRL 1564
Query: 386 EQRVRELESELD---AEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQ 556
+ + +++SE+D AE+ ++ K R R ++ + D + +++ L +K++
Sbjct: 1565 QLELNQIKSEIDRKIAEKDEEIDQLK--RNHLRIVESMQSTLDAEIRSRNEALRLKKKME 1622
Query: 557 GKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLR 703
G + + Q+ A +AA R Q ++D++ D A + + L+
Sbjct: 1623 GDLNEMEIQLSHANRMAAEAQKNLRNTQGTLKDTQIHLDDALRTQEDLK 1671
Score = 63.2 bits (152), Expect = 1e-09
Identities = 52/235 (22%), Positives = 110/235 (46%), Gaps = 14/235 (5%)
Frame = +2
Query: 17 ENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQAN 196
E+ E ++K +L D +EL T K+E + A ++ ++ + E +
Sbjct: 930 EDEEEINAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAVLD 989
Query: 197 E------QAKKAVADS-SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAM 355
E + KKA+ ++ + D+++ E++ + K K +LE Q +L+ L++ + M
Sbjct: 990 ETIAKLTKEKKALQEAHQQTLDDLQVEEDKVNTLTKAKTKLEQQVDDLEGSLEQEKKLRM 1049
Query: 356 XXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQ 535
E+ R+LE +L ++ + + ++++ +LK+ + + + E Q
Sbjct: 1050 DL--------ERAKRKLEGDLKLAHDSIMDLENDKQQLDEKLKKKDFEISQIQSKIEDEQ 1101
Query: 536 DLVEKLQGKIKTYKRQVE------EAEEIAAVNLAKYR-KIQQEIEDSEERADQA 679
L +LQ KIK + ++E EAE + K+R + +E+E+ ER ++A
Sbjct: 1102 ALGMQLQKKIKELQARIEELEEEIEAERTSRAKAEKHRADLSRELEEISERLEEA 1156
Score = 48.5 bits (114), Expect = 3e-05
Identities = 55/286 (19%), Positives = 113/286 (39%), Gaps = 49/286 (17%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQ- 190
++N +R ++ E EK EL +++L+ S K LE ++ L E + Q
Sbjct: 1210 IDNLQRVKQKLEKEKSELKMEIDDLASNMESVSKAKANLEKMCRTLEDQLSEIKTKEEQN 1269
Query: 191 -----------------ANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNK-- 313
E +++A + + R +Q QQI+++K+ LE + K
Sbjct: 1270 QRMINDLNTQRARLQTETGEYSRQAEEKDALISQLSRGKQGFTQQIEELKRHLEEEIKAK 1329
Query: 314 --------------ELQAKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQ-KRHVET 448
EL + E E A V + ++ + + +R E
Sbjct: 1330 NALAHALQSARHDCELLREQYEEEQEAKGELQRALSKANSEVAQWRTKYETDAIQRTEEL 1389
Query: 449 QKNTRKIERRLKE-------IGLQTDEDKKNQERLQDLVEKLQGKIK-------TYKRQV 586
++ +K+ +RL++ + + +K ++RLQ+ VE L ++ ++
Sbjct: 1390 EEAKKKLAQRLQDAEEHVEAVNAKCASLEKTKQRLQNEVEDLMVDVERSNAACAALDKKQ 1449
Query: 587 EEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSV 724
+ ++I A KY + Q E+E S++ + L K++ S+
Sbjct: 1450 KNFDKILAEWKQKYEETQTELEASQKESRSLSTELFKMKNAYEESL 1495
Score = 39.3 bits (90), Expect = 0.017
Identities = 25/126 (19%), Positives = 59/126 (46%), Gaps = 14/126 (11%)
Frame = +2
Query: 386 EQRVRELESELDAEQKRHVETQKNTRKIERRLK-----------EIGLQTDEDKKNQERL 532
E+ + ++ E + ++ +++ +++E ++ ++ + D +ER
Sbjct: 849 EKEMANMKEEFEKTKEELAKSEAKRKELEEKMVVLLQEKNDLQLQVQAEADSLADAEERC 908
Query: 533 QDLVE---KLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLR 703
L++ +L+ KIK + E+ EEI A AK RK++ E + ++ D E L K+
Sbjct: 909 DQLIKTKIQLEAKIKEVTERAEDEEEINAELTAKKRKLEDECSELKKDIDDLELTLAKVE 968
Query: 704 TKNRSS 721
+ ++
Sbjct: 969 KEKHAT 974
>sp|Q5SX40|MYH1_MOUSE Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) Length = 1942 Score = 229 bits (584), Expect = 9e-60 Identities = 117/238 (49%), Positives = 171/238 (71%) Frame = +2 Query: 2 VRGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNE 181 +R +LE +ER+RK E E ++ ++R+ L Q++S + TK+KLE D++ +Q ++E+ E Sbjct: 1695 LRATLEQTERSRKIAEQELLDASERVQLLHTQNTSLINTKKKLETDISQIQGEMEDIVQE 1754 Query: 182 ARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXX 361 AR A E+AKKA+ D++ + +E+++EQ+ + ++++KK LE K+LQ +LDE+E A+ Sbjct: 1755 ARNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNLEQTVKDLQHRLDEAEQLALKG 1814 Query: 362 XXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDL 541 E RVRELE E++ EQKR+VE K RK ERR+KE+ QT+ED+KN RLQDL Sbjct: 1815 GKKQIQKLEARVRELEGEVENEQKRNVEAIKGLRKHERRVKELTYQTEEDRKNVLRLQDL 1874 Query: 542 VEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNR 715 V+KLQ K+K YKRQ EEAEE + VNLAK+RKIQ E+E++EERAD AE + KLR K+R Sbjct: 1875 VDKLQSKVKAYKRQAEEAEEQSNVNLAKFRKIQHELEEAEERADIAESQVNKLRVKSR 1932
Score = 71.6 bits (174), Expect = 3e-12
Identities = 50/227 (22%), Positives = 114/227 (50%)
Frame = +2
Query: 47 ESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQAKKAVADS 226
E++ E+T+R + ++ A KRKLE + + ++ D+++ + ++
Sbjct: 923 EAKIKEVTERAEDEEEINAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKV 982
Query: 227 SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXXEQRVREL 406
L +E+ E ++ K KK L+ +++ L ++E + + EQ+V +L
Sbjct: 983 KNLTEEMAGLDETIAKLTKEKKALQEAHQQTLDDL-QAEEDKVNTLTKAKIKLEQQVDDL 1041
Query: 407 ESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQV 586
E L+ E+K ++ ++ RK+E LK T + + ++++L + ++K + ++ + ++
Sbjct: 1042 EGSLEQEKKIRMDLERAKRKLEGDLKLAQESTMDVENDKQQLDEKLKKKEFEMSNLQSKI 1101
Query: 587 EEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVS 727
E+ + + K +++Q IE+ EE +AE+A + K RS +S
Sbjct: 1102 EDEQALGMQLQKKIKELQARIEELEEEI-EAERASRAKAEKQRSDLS 1147
Score = 71.6 bits (174), Expect = 3e-12
Identities = 63/256 (24%), Positives = 118/256 (46%), Gaps = 12/256 (4%)
Frame = +2
Query: 5 RGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEAS--N 178
R S +E+ R + E E+++RL E +S+ + +K EA+ M+ DLEEA+ +
Sbjct: 1133 RASRAKAEKQRSDLSRELEEISERLEEAGGATSAQIEMNKKREAEFQKMRRDLEEATLQH 1192
Query: 179 EARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMX 358
EA A + K A D + + E + ++K++LE + E++ ++D+ +N M
Sbjct: 1193 EATAATLRKKHA--------DSVAELGEQIDNLQRVKQKLEKEKSEMKMEIDDLASN-ME 1243
Query: 359 XXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQD 538
E+ R LE ++ + + E Q+ ++ + + ++ E + +
Sbjct: 1244 VISKSKGNLEKMCRTLEDQVSELKTKEEEQQRLINELTAQRGRLQTESGEYSRQLDEKDS 1303
Query: 539 LVEKL-QGK------IKTYKRQVEE---AEEIAAVNLAKYRKIQQEIEDSEERADQAEQA 688
LV +L +GK I+ KRQ+EE A+ A L R + + E +A+
Sbjct: 1304 LVSQLSRGKQAFTQQIEELKRQLEEEIKAKSALAHALQSSRHDCDLLREQYEEEQEAKAE 1363
Query: 689 LQKLRTKNRSSVSTAR 736
LQ+ +K S V+ R
Sbjct: 1364 LQRAMSKANSEVAQWR 1379
Score = 69.3 bits (168), Expect = 1e-11
Identities = 54/242 (22%), Positives = 113/242 (46%), Gaps = 1/242 (0%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
LE +R KN + E +LT+++ E + K+++E + + +Q+ LEEA
Sbjct: 1502 LETLKRENKNLQQEISDLTEQIAEGGKRIHELEKIKKQIEQEKSELQAALEEAEASLEHE 1561
Query: 194 NEQAKKAVADSSRLFDEI-RQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXX 370
+ + + +++ EI R+ E ++ID++K+ + +Q+ LD +E +
Sbjct: 1562 EGKILRIQLELNQVKSEIDRKIAEKDEEIDQLKRNHIRVVESMQSTLD-AEIRSRNDAIR 1620
Query: 371 XXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEK 550
E + E+E +L+ + E +N R + LK+ L D+ + QE L++ +
Sbjct: 1621 LKKKMEGDLNEMEIQLNHSNRMAAEALRNYRNTQGILKDTQLHLDDALRGQEDLKEQLAM 1680
Query: 551 LQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVST 730
++ + + ++EE + +QE+ D+ ER +Q L T+N S ++T
Sbjct: 1681 VERRANLLQAEIEELRATLEQTERSRKIAEQELLDASER-------VQLLHTQNTSLINT 1733
Query: 731 AR 736
+
Sbjct: 1734 KK 1735
Score = 67.0 bits (162), Expect = 7e-11
Identities = 48/229 (20%), Positives = 108/229 (47%), Gaps = 3/229 (1%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
++ ++N + E + E + + R L +L +++ EE+ + +
Sbjct: 1450 DKKQRNFDKILAEWKQKYEETHAELEASQKESRSLSTELFKIKNAYEESLDHLETLKREN 1509
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXX 385
K + S L ++I + + +++KIKKQ+E + ELQA L+E+E ++
Sbjct: 1510 KNLQQEISDLTEQIAEGGKRIHELEKIKKQIEQEKSELQAALEEAE-ASLEHEEGKILRI 1568
Query: 386 EQRVRELESELD---AEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQ 556
+ + +++SE+D AE+ ++ K R R ++ + D + +++ L +K++
Sbjct: 1569 QLELNQVKSEIDRKIAEKDEEIDQLK--RNHIRVVESMQSTLDAEIRSRNDAIRLKKKME 1626
Query: 557 GKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLR 703
G + + Q+ + +AA L YR Q ++D++ D A + + L+
Sbjct: 1627 GDLNEMEIQLNHSNRMAAEALRNYRNTQGILKDTQLHLDDALRGQEDLK 1675
Score = 65.9 bits (159), Expect = 2e-10
Identities = 50/234 (21%), Positives = 108/234 (46%), Gaps = 7/234 (2%)
Frame = +2
Query: 17 ENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQAN 196
E+ E ++K +L D +EL T K+E + A ++ ++ + E +
Sbjct: 934 EDEEEINAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAGLD 993
Query: 197 E------QAKKAVADS-SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAM 355
E + KKA+ ++ + D+++ E++ + K K +LE Q +L+ L++ + M
Sbjct: 994 ETIAKLTKEKKALQEAHQQTLDDLQAEEDKVNTLTKAKIKLEQQVDDLEGSLEQEKKIRM 1053
Query: 356 XXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQ 535
E+ R+LE +L Q+ ++ + + ++++ +LK+ + + E Q
Sbjct: 1054 DL--------ERAKRKLEGDLKLAQESTMDVENDKQQLDEKLKKKEFEMSNLQSKIEDEQ 1105
Query: 536 DLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQK 697
L +LQ KIK + ++EE EE A K +++ D ++ + L++
Sbjct: 1106 ALGMQLQKKIKELQARIEELEEEIEAERASRAKAEKQRSDLSRELEEISERLEE 1159
Score = 45.8 bits (107), Expect = 2e-04
Identities = 49/221 (22%), Positives = 105/221 (47%), Gaps = 28/221 (12%)
Frame = +2
Query: 119 KRKLEADLAAMQSDLEEASNEARQA--NEQAKKAVADSSRLF--DEIRQEQEHAQQIDKI 286
K+K+E DL M+ L ++ A +A N + + + ++L D +R +++ +Q+ +
Sbjct: 1622 KKKMEGDLNEMEIQLNHSNRMAAEALRNYRNTQGILKDTQLHLDDALRGQEDLKEQLAMV 1681
Query: 287 KKQ---LEAQNKELQAKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKN 457
+++ L+A+ +EL+A L+++E + +++ E E LDA ++ + +N
Sbjct: 1682 ERRANLLQAEIEELRATLEQTERS-------------RKIAEQEL-LDASERVQLLHTQN 1727
Query: 458 TRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIA---------- 607
T I + K L+TD + ++QG+++ ++ AEE A
Sbjct: 1728 TSLINTKKK---LETD------------ISQIQGEMEDIVQEARNAEEKAKKAITDAAMM 1772
Query: 608 ----------AVNLAKYRK-IQQEIEDSEERADQAEQALQK 697
+ +L + +K ++Q ++D + R D+AEQ K
Sbjct: 1773 AEELKKEQDTSAHLERMKKNLEQTVKDLQHRLDEAEQLALK 1813
Score = 44.3 bits (103), Expect = 5e-04
Identities = 54/286 (18%), Positives = 114/286 (39%), Gaps = 49/286 (17%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLE----------ADLAAMQSDL 163
++N +R ++ E EK E+ +++L+ +K LE ++L + +
Sbjct: 1214 IDNLQRVKQKLEKEKSEMKMEIDDLASNMEVISKSKGNLEKMCRTLEDQVSELKTKEEEQ 1273
Query: 164 EEASNEARQANEQAKKAVADSSRLFDE--------IRQEQEHAQQIDKIKKQLEAQNK-- 313
+ NE + + + SR DE R +Q QQI+++K+QLE + K
Sbjct: 1274 QRLINELTAQRGRLQTESGEYSRQLDEKDSLVSQLSRGKQAFTQQIEELKRQLEEEIKAK 1333
Query: 314 --------------ELQAKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQ-KRHVET 448
+L + E E A V + ++ + + +R E
Sbjct: 1334 SALAHALQSSRHDCDLLREQYEEEQEAKAELQRAMSKANSEVAQWRTKYETDAIQRTEEL 1393
Query: 449 QKNTRKIERRLKE-------IGLQTDEDKKNQERLQDLVEKLQGKIK-------TYKRQV 586
++ +K+ +RL++ + + +K ++RLQ+ VE L ++ ++
Sbjct: 1394 EEAKKKLAQRLQDAEEHVEAVNAKCASLEKTKQRLQNEVEDLMIDVERTNAACAALDKKQ 1453
Query: 587 EEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSV 724
++I A KY + E+E S++ + L K++ S+
Sbjct: 1454 RNFDKILAEWKQKYEETHAELEASQKESRSLSTELFKIKNAYEESL 1499
Score = 39.3 bits (90), Expect = 0.017
Identities = 26/126 (20%), Positives = 60/126 (47%), Gaps = 14/126 (11%)
Frame = +2
Query: 386 EQRVRELESELDAEQKRHVETQKNTRKIERRL-------KEIGLQTDEDKKN----QERL 532
E+ + ++ E + ++ + + +++E ++ ++ LQ + + +ER
Sbjct: 853 EKEMANMKEEFEKAKENLAKAEAKRKELEEKMVALMQEKNDLQLQVQSEADSLADAEERC 912
Query: 533 QDLVE---KLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLR 703
L++ +L+ KIK + E+ EEI A AK RK++ E + ++ D E L K+
Sbjct: 913 DQLIKTKIQLEAKIKEVTERAEDEEEINAELTAKKRKLEDECSELKKDIDDLELTLAKVE 972
Query: 704 TKNRSS 721
+ ++
Sbjct: 973 KEKHAT 978
>sp|P13533|MYH6_HUMAN Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha) Length = 1939 Score = 229 bits (584), Expect = 9e-60 Identities = 117/238 (49%), Positives = 170/238 (71%) Frame = +2 Query: 2 VRGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNE 181 +R +E +ER+RK E E +E ++R+ L Q++S + K+K+EADL +QS++EEA E Sbjct: 1690 LRAVVEQTERSRKLAEQELIETSERVQLLHSQNTSLINQKKKMEADLTQLQSEVEEAVQE 1749 Query: 182 ARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXX 361 R A E+AKKA+ D++ + +E+++EQ+ + ++++KK +E K+LQ +LDE+E A+ Sbjct: 1750 CRNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNMEQTIKDLQHRLDEAEQIALKG 1809 Query: 362 XXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDL 541 E RVRELE EL+AEQKR+ E+ K RK ERR+KE+ QT+EDKKN RLQDL Sbjct: 1810 GKKQLQKLEARVRELEGELEAEQKRNAESVKGMRKSERRIKELTYQTEEDKKNLLRLQDL 1869 Query: 542 VEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNR 715 V+KLQ K+K YKRQ EEAEE A NL+K+RK+Q E++++EERAD AE + KLR K+R Sbjct: 1870 VDKLQLKVKAYKRQAEEAEEQANTNLSKFRKVQHELDEAEERADIAESQVNKLRAKSR 1927
Score = 68.6 bits (166), Expect = 3e-11
Identities = 55/236 (23%), Positives = 111/236 (47%), Gaps = 14/236 (5%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
LE+ E ++K +L D +EL T K+E + A ++ ++ + E
Sbjct: 928 LEDEEEMNAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAGL 987
Query: 194 NE------QAKKAVADS-SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNA 352
+E + KKA+ ++ + D+++ E++ + K K +LE Q +L+ L++ +
Sbjct: 988 DEIIAKLTKEKKALQEAHQQALDDLQVEEDKVNSLSKSKVKLEQQVDDLEGSLEQEKKVR 1047
Query: 353 MXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERL 532
M E+ R+LE +L Q+ ++ + + ++E +LK+ ++ E
Sbjct: 1048 MDL--------ERAKRKLEGDLKLTQESIMDLENDKLQLEEKLKKKEFDINQQNSKIEDE 1099
Query: 533 QDLVEKLQGKIKTYKRQVE------EAEEIAAVNLAKYRK-IQQEIEDSEERADQA 679
Q L +LQ K+K + ++E EAE A + K R + +E+E+ ER ++A
Sbjct: 1100 QALALQLQKKLKENQARIEELEEELEAERTARAKVEKLRSDLSRELEEISERLEEA 1155
Score = 63.9 bits (154), Expect = 6e-10
Identities = 59/247 (23%), Positives = 112/247 (45%), Gaps = 10/247 (4%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
E+ R + E E+++RL E +S + +K EA+ M+ DLEEA+ +
Sbjct: 1135 EKLRSDLSRELEEISERLEEAGGATSVQIEMNKKREAEFQKMRRDLEEATLQHEATAAAL 1194
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXX 385
+K ADS + + E + ++K++LE + E + +LD+ +N M
Sbjct: 1195 RKKHADS------VAELGEQIDNLQRVKQKLEKEKSEFKLELDDVTSN-MEQIIKAKANL 1247
Query: 386 EQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKL-QGK 562
E+ R LE + + + + E Q++ + ++ + E + E + L+ +L +GK
Sbjct: 1248 EKVSRTLEDQANEYRVKLEEAQRSLNDFTTQRAKLQTENGELARQLEEKEALISQLTRGK 1307
Query: 563 I------KTYKRQVEE---AEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNR 715
+ + KRQ+EE A+ A L R + + E +A+ LQ++ +K
Sbjct: 1308 LSYTQQMEDLKRQLEEEGKAKNALAHALQSARHDCDLLREQYEEETEAKAELQRVLSKAN 1367
Query: 716 SSVSTAR 736
S V+ R
Sbjct: 1368 SEVAQWR 1374
Score = 63.9 bits (154), Expect = 6e-10
Identities = 44/228 (19%), Positives = 109/228 (47%), Gaps = 2/228 (0%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
++ ++N + E + E + S R L +L +++ EE+ +
Sbjct: 1445 DKKQRNFDKILAEWKQKYEESQSELESSQKEARSLSTELFKLKNAYEESLEHLETFKREN 1504
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXX 385
K + S L +++ + ++ +++K++KQLE + ELQ+ L+E+E ++
Sbjct: 1505 KNLQEEISDLTEQLGEGGKNVHELEKVRKQLEVEKLELQSALEEAE-ASLEHEEGKILRA 1563
Query: 386 EQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDL--VEKLQG 559
+ ++++E++ + E + ++ +R+ + LQT D + + R + L +K++G
Sbjct: 1564 QLEFNQIKAEIERKLAEKDEEMEQAKRNHQRVVD-SLQTSLDAETRSRNEVLRVKKKMEG 1622
Query: 560 KIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLR 703
+ + Q+ A +AA + + +Q ++D++ + D A +A L+
Sbjct: 1623 DLNEMEIQLSHANRMAAEAQKQVKSLQSLLKDTQIQLDDAVRANDDLK 1670
Score = 52.0 bits (123), Expect = 2e-06
Identities = 53/259 (20%), Positives = 108/259 (41%), Gaps = 18/259 (6%)
Frame = +2
Query: 2 VRGSLENSERNRKNTESEKVELTDRLNELSVQSSS--------------FMATKRKLEAD 139
++ +LE SE RK E + V L N+L +Q + + K +LEA
Sbjct: 861 IKETLEKSEARRKELEEKMVSLLQEKNDLQLQVQAEQDNLNDAEERCDQLIKNKIQLEAK 920
Query: 140 LAAMQSDLEEASNEARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKEL 319
+ M LE+ + + +K + S L +I + +++K K E + K L
Sbjct: 921 VKEMNERLEDEEEMNAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKVKNL 980
Query: 320 QAKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQ 499
++ + E + L+ +L E+ + K+ K+E+++ ++
Sbjct: 981 TEEMAGLDEIIAKLTKEKKALQEAHQQALD-DLQVEEDKVNSLSKSKVKLEQQVDDLEGS 1039
Query: 500 TDEDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQA 679
+++KK + L+ KL+G +K + + + E K +K + +I + +
Sbjct: 1040 LEQEKKVRMDLERAKRKLEGDLKLTQESIMDLENDKLQLEEKLKKKEFDINQQNSKIED- 1098
Query: 680 EQA----LQKLRTKNRSSV 724
EQA LQK +N++ +
Sbjct: 1099 EQALALQLQKKLKENQARI 1117
Score = 48.1 bits (113), Expect = 4e-05
Identities = 58/291 (19%), Positives = 117/291 (40%), Gaps = 54/291 (18%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
++N +R ++ E EK E L++++ + K +A+L + LE+ +NE R
Sbjct: 1209 IDNLQRVKQKLEKEKSEFKLELDDVTSNMEQII----KAKANLEKVSRTLEDQANEYRVK 1264
Query: 194 NEQAKKAVADSSRLFDEIRQEQ----------------------EHAQQIDKIKKQLEAQ 307
E+A++++ D + +++ E + QQ++ +K+QLE +
Sbjct: 1265 LEEAQRSLNDFTTQRAKLQTENGELARQLEEKEALISQLTRGKLSYTQQMEDLKRQLEEE 1324
Query: 308 NKELQAKL------------------DESENNAMXXXXXXXXXXEQRVRELESELDAEQK 433
K A +E+E A E + E DA Q+
Sbjct: 1325 GKAKNALAHALQSARHDCDLLREQYEEETEAKAELQRVLSKANSEVAQWRTKYETDAIQR 1384
Query: 434 RHVETQKNTRKIERRLKE-------IGLQTDEDKKNQERLQDLVEKLQGKIK-------T 571
E ++ +K+ +RL++ + + +K + RLQ+ +E L ++
Sbjct: 1385 TE-ELEEAKKKLAQRLQDAEEAVEAVNAKCSSLEKTKHRLQNEIEDLMVDVERSNAAAAA 1443
Query: 572 YKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSV 724
++ ++I A KY + Q E+E S++ A L KL+ S+
Sbjct: 1444 LDKKQRNFDKILAEWKQKYEESQSELESSQKEARSLSTELFKLKNAYEESL 1494
Score = 42.4 bits (98), Expect = 0.002
Identities = 42/222 (18%), Positives = 91/222 (40%), Gaps = 5/222 (2%)
Frame = +2
Query: 89 SVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQAKKAVADSSRLFDEIRQEQEHA 268
S ++ MAT ++ + ++ LE++ ++ E+ + + + L +++ EQ++
Sbjct: 844 SAETEKEMATMKE---EFGRIKETLEKSEARRKELEEKMVSLLQEKNDLQLQVQAEQDNL 900
Query: 269 QQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQKRHVET 448
++ QL +L+AK V+E+ L+ E++ + E
Sbjct: 901 NDAEERCDQLIKNKIQLEAK----------------------VKEMNERLEDEEEMNAEL 938
Query: 449 QKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKY 628
RK+E E+ D+ + +++ + K+K ++ +EI A L K
Sbjct: 939 TAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAGLDEIIA-KLTKE 997
Query: 629 RKI-----QQEIEDSEERADQAEQALQKLRTKNRSSVSTARG 739
+K QQ ++D + D+ +L K + K V G
Sbjct: 998 KKALQEAHQQALDDLQVEEDKV-NSLSKSKVKLEQQVDDLEG 1038
>sp|P13539|MYH6_MESAU Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha) Length = 1939 Score = 229 bits (583), Expect = 1e-59 Identities = 116/238 (48%), Positives = 171/238 (71%) Frame = +2 Query: 2 VRGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNE 181 +R +E +ER+RK E E +E ++R+ L Q++S + K+K+EADL +Q+++EEA E Sbjct: 1690 LRAVVEQTERSRKLAEQELIETSERVQLLHSQNTSLINQKKKMEADLTQLQTEVEEAVQE 1749 Query: 182 ARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXX 361 R A E+AKKA+ D++ + +E+++EQ+ + ++++KK +E K+LQ +LDE+E A+ Sbjct: 1750 CRNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNMEQTIKDLQHRLDEAEQIALKG 1809 Query: 362 XXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDL 541 E RVRELE+EL+AEQKR+ E+ K RK ERR+KE+ QT+EDKKN RLQDL Sbjct: 1810 GKKQLQKLEARVRELENELEAEQKRNAESVKGMRKSERRIKELTYQTEEDKKNLVRLQDL 1869 Query: 542 VEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNR 715 V+KLQ K+K YKRQ EEAEE A NL+K+RK+Q E++++EERAD AE + KLR K+R Sbjct: 1870 VDKLQLKVKAYKRQAEEAEEQANTNLSKFRKVQHELDEAEERADIAESQVNKLRAKSR 1927
Score = 69.7 bits (169), Expect = 1e-11
Identities = 56/236 (23%), Positives = 111/236 (47%), Gaps = 14/236 (5%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
LE+ E S+K +L D +EL T K+E + A ++ ++ + E
Sbjct: 928 LEDEEEMNAELTSKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAGL 987
Query: 194 NE------QAKKAVADS-SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNA 352
+E + KKA+ ++ + D+++ E++ + K K +LE Q +L+ L++ +
Sbjct: 988 DEIIAKLTKEKKALQEAHQQALDDLQAEEDKVNTLTKSKVKLEQQVDDLEGSLEQEKKVR 1047
Query: 353 MXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERL 532
M E+ R+LE +L+ Q+ ++ + + ++E +LK+ + E
Sbjct: 1048 MDL--------ERAKRKLEGDLNVTQESIMDLENDKLQLEEKLKKKEFDISQQNSKIEDE 1099
Query: 533 QDLVEKLQGKIKTYKRQVE------EAEEIAAVNLAKYRK-IQQEIEDSEERADQA 679
Q L +LQ K+K + ++E EAE A + K R + +E+E+ ER ++A
Sbjct: 1100 QALALQLQKKLKENQARIEELEEELEAERTARAKVEKLRSDLTRELEEISERLEEA 1155
Score = 63.2 bits (152), Expect = 1e-09
Identities = 59/247 (23%), Positives = 113/247 (45%), Gaps = 10/247 (4%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
E+ R + E E+++RL E +S + +K EA+ M+ DLEEA+ +
Sbjct: 1135 EKLRSDLTRELEEISERLEEAGGATSVQIEMNKKREAEFQKMRRDLEEATLQHEATAAAL 1194
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXX 385
+K ADS + + E + ++K++LE + E + +LD+ +N M
Sbjct: 1195 RKKHADS------VAELGEQIDNLQRVKQKLEKEKSEFKLELDDVTSN-MEQIIKAKANL 1247
Query: 386 EQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKL-QGK 562
E+ R LE + + + + E+Q++ + ++ + E + E + L+ +L +GK
Sbjct: 1248 EKVSRTLEDQANEYRVKLEESQRSLNDFTTQRAKLQTENGELARQLEEKEALISQLTRGK 1307
Query: 563 I------KTYKRQVEE---AEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNR 715
+ + KRQ+EE A+ A L R + + E +A+ LQ++ +K
Sbjct: 1308 LSYTQQMEDLKRQLEEEGKAKNALAHALQSARHDCDLLREQYEEEMEAKAELQRVLSKAN 1367
Query: 716 SSVSTAR 736
S V+ R
Sbjct: 1368 SEVAQWR 1374
Score = 62.4 bits (150), Expect = 2e-09
Identities = 44/228 (19%), Positives = 106/228 (46%), Gaps = 2/228 (0%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
++ ++N + E + E + S R L +L +++ EE+ +
Sbjct: 1445 DKKQRNFDKILAEWKQKYEESQSELESSQKEARSLSTELFKLKNAYEESLEHLETFKREN 1504
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXX 385
K + S L +++ + ++ +++K++KQLE + ELQ+ L+E+E ++
Sbjct: 1505 KNLQEEISDLTEQLGEGGKNVHELEKVRKQLEVEKMELQSALEEAE-ASLEHEEGKILRA 1563
Query: 386 EQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDL--VEKLQG 559
+ ++++E++ + E + ++ R+ + LQT D + + R + L +K++G
Sbjct: 1564 QLEFNQIKAEIERKLAEKDEEMEQAKRNHLRVVD-SLQTSLDAETRSRNEALRVKKKMEG 1622
Query: 560 KIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLR 703
+ + Q+ +A IA+ + Q ++D++ + D A A L+
Sbjct: 1623 DLNEMEIQLSQANRIASEAQKHLKNAQAHLKDTQLQLDDALHANDDLK 1670
Score = 52.8 bits (125), Expect = 1e-06
Identities = 55/259 (21%), Positives = 107/259 (41%), Gaps = 18/259 (6%)
Frame = +2
Query: 2 VRGSLENSERNRKNTESEKVELT--------------DRLNELSVQSSSFMATKRKLEAD 139
V+ SLE SE RK E + V L D LN+ + + K +LEA
Sbjct: 861 VKESLEKSEARRKELEEKMVSLLQEKNDLQFQVQAEQDNLNDAEERCDQLIKNKIQLEAK 920
Query: 140 LAAMQSDLEEASNEARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKEL 319
+ M LE+ + + +K + S L +I + +++K K E + K L
Sbjct: 921 VKEMTERLEDEEEMNAELTSKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKVKNL 980
Query: 320 QAKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQ 499
++ + E + L+ +L AE+ + K+ K+E+++ ++
Sbjct: 981 TEEMAGLDEIIAKLTKEKKALQEAHQQALD-DLQAEEDKVNTLTKSKVKLEQQVDDLEGS 1039
Query: 500 TDEDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQA 679
+++KK + L+ KL+G + + + + E K +K + +I + +
Sbjct: 1040 LEQEKKVRMDLERAKRKLEGDLNVTQESIMDLENDKLQLEEKLKKKEFDISQQNSKIED- 1098
Query: 680 EQA----LQKLRTKNRSSV 724
EQA LQK +N++ +
Sbjct: 1099 EQALALQLQKKLKENQARI 1117
Score = 46.2 bits (108), Expect = 1e-04
Identities = 57/291 (19%), Positives = 116/291 (39%), Gaps = 54/291 (18%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
++N +R ++ E EK E L++++ + K +A+L + LE+ +NE R
Sbjct: 1209 IDNLQRVKQKLEKEKSEFKLELDDVTSNMEQII----KAKANLEKVSRTLEDQANEYRVK 1264
Query: 194 NEQAKKAVADSSRLFDEIRQEQ----------------------EHAQQIDKIKKQLEAQ 307
E++++++ D + +++ E + QQ++ +K+QLE +
Sbjct: 1265 LEESQRSLNDFTTQRAKLQTENGELARQLEEKEALISQLTRGKLSYTQQMEDLKRQLEEE 1324
Query: 308 NKELQAKL------------------DESENNAMXXXXXXXXXXEQRVRELESELDAEQK 433
K A +E E A E + E DA Q+
Sbjct: 1325 GKAKNALAHALQSARHDCDLLREQYEEEMEAKAELQRVLSKANSEVAQWRTKYETDAIQR 1384
Query: 434 RHVETQKNTRKIERRLKE-------IGLQTDEDKKNQERLQDLVEKLQGKIK-------T 571
E ++ +K+ +RL++ + + +K + RLQ+ +E L ++
Sbjct: 1385 TE-ELEEAKKKLAQRLQDAEEAVEAVNAKCSSLEKTKHRLQNEIEDLMVDVERSNAAAAA 1443
Query: 572 YKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSV 724
++ ++I A KY + Q E+E S++ A L KL+ S+
Sbjct: 1444 LDKKQRNFDKILAEWKQKYEESQSELESSQKEARSLSTELFKLKNAYEESL 1494
Score = 43.9 bits (102), Expect = 7e-04
Identities = 40/190 (21%), Positives = 79/190 (41%), Gaps = 2/190 (1%)
Frame = +2
Query: 161 LEEASNEARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKI--KKQLEAQNKELQAKLD 334
L+ A E AN + + S E R+++ + + + K L+ Q + Q L+
Sbjct: 842 LKSAETEKEMANMKEEFGRVKESLEKSEARRKELEEKMVSLLQEKNDLQFQVQAEQDNLN 901
Query: 335 ESENNAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDK 514
++E E +V+E+ L+ E++ + E RK+E E+ D+ +
Sbjct: 902 DAEERC-DQLIKNKIQLEAKVKEMTERLEDEEEMNAELTSKKRKLEDECSELKKDIDDLE 960
Query: 515 KNQERLQDLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQ 694
+++ + K+K ++ +EI A L K +K QE ++ D +
Sbjct: 961 LTLAKVEKEKHATENKVKNLTEEMAGLDEIIA-KLTKEKKALQEAH--QQALDDLQAEED 1017
Query: 695 KLRTKNRSSV 724
K+ T +S V
Sbjct: 1018 KVNTLTKSKV 1027
Score = 40.8 bits (94), Expect = 0.006
Identities = 33/164 (20%), Positives = 74/164 (45%), Gaps = 8/164 (4%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
ER +KN E +L RL+E + + K++L+ L A +LE ++ N ++
Sbjct: 1782 ERMKKNMEQTIKDLQHRLDE--AEQIALKGGKKQLQK-LEARVRELENELEAEQKRNAES 1838
Query: 206 KKAVADSSRLFDEIRQEQEHAQQ--------IDKIKKQLEAQNKELQAKLDESENNAMXX 361
K + S R E+ + E ++ +DK++ +++A ++ + +++ N
Sbjct: 1839 VKGMRKSERRIKELTYQTEEDKKNLVRLQDLVDKLQLKVKAYKRQAEEAEEQANTNL--- 1895
Query: 362 XXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIG 493
+ R+++ ELD ++R + K+ + ++IG
Sbjct: 1896 ---------SKFRKVQHELDEAEERADIAESQVNKLRAKSRDIG 1930
>sp|Q9UKX2|MYH2_HUMAN Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa) Length = 1941 Score = 228 bits (581), Expect = 2e-59 Identities = 114/238 (47%), Positives = 171/238 (71%) Frame = +2 Query: 2 VRGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNE 181 +R +LE +ER+RK E E ++ ++R+ L Q++S + TK+KLE D++ MQ ++E+ E Sbjct: 1694 LRATLEQTERSRKIAEQELLDASERVQLLHTQNTSLINTKKKLETDISQMQGEMEDILQE 1753 Query: 182 ARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXX 361 AR A E+AKKA+ D++ + +E+++EQ+ + ++++KK +E K+LQ +LDE+E A+ Sbjct: 1754 ARNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNMEQTVKDLQLRLDEAEQLALKG 1813 Query: 362 XXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDL 541 E RVRELE E+++EQKR+ E K RK ERR+KE+ QT+ED+KN RLQDL Sbjct: 1814 GKKQIQKLEARVRELEGEVESEQKRNAEAVKGLRKHERRVKELTYQTEEDRKNILRLQDL 1873 Query: 542 VEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNR 715 V+KLQ K+K+YKRQ EEAEE + NLAK+RK+Q E+E++EERAD AE + KLR K+R Sbjct: 1874 VDKLQAKVKSYKRQAEEAEEQSNTNLAKFRKLQHELEEAEERADIAESQVNKLRVKSR 1931
Score = 72.0 bits (175), Expect = 2e-12
Identities = 57/251 (22%), Positives = 116/251 (46%), Gaps = 16/251 (6%)
Frame = +2
Query: 5 RGSLENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEAS--N 178
R S +E+ R + E E+++RL E +S+ + +K EA+ M+ DLEEA+ +
Sbjct: 1132 RASRAKAEKQRSDLSRELEEISERLEEAGGATSAQIEMNKKREAEFQKMRRDLEEATLQH 1191
Query: 179 EARQANEQAKKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMX 358
EA A + K A D + + E + ++K++LE + E++ ++D+ +N +
Sbjct: 1192 EATAATLRKKHA--------DSVAELGEQIDNLQRVKQKLEKEKSEMKMEIDDLASN-VE 1242
Query: 359 XXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQD 538
E+ R LE +L + + E Q+ + + + ++ E + + +
Sbjct: 1243 TVSKAKGNLEKMCRTLEDQLSELKSKEEEQQRLINDLTAQRGRLQTESGEFSRQLDEKEA 1302
Query: 539 LVEKLQGKIKTYKRQVEE-----AEEIAAVNLAKYR---------KIQQEIEDSEERADQ 676
LV +L + + +Q+EE EEI A N + ++++ E+ +E +
Sbjct: 1303 LVSQLSRGKQAFTQQIEELKRQLEEEIKAKNALAHALQSSRHDCDLLREQYEEEQESKAE 1362
Query: 677 AEQALQKLRTK 709
++AL K T+
Sbjct: 1363 LQRALSKANTE 1373
Score = 70.5 bits (171), Expect = 7e-12
Identities = 50/227 (22%), Positives = 112/227 (49%)
Frame = +2
Query: 47 ESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQAKKAVADS 226
E++ E+T+R + ++ A KRKLE + + ++ D+++ + ++
Sbjct: 922 EAKIKEVTERAEDEEEINAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKV 981
Query: 227 SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXXEQRVREL 406
L +E+ E ++ K KK L+ +++ L ++E + + EQ+V +L
Sbjct: 982 KNLTEEMAGLDETIAKLTKEKKALQEAHQQTLDDL-QAEEDKVNTLTKAKIKLEQQVDDL 1040
Query: 407 ESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQV 586
E L+ E+K ++ ++ RK+E LK + + +++L + ++K + +I + ++
Sbjct: 1041 EGSLEQEKKLRMDLERAKRKLEGDLKLAQESIMDIENEKQQLDEKLKKKEFEISNLQSKI 1100
Query: 587 EEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVS 727
E+ + + K +++Q IE+ EE +AE+A + K RS +S
Sbjct: 1101 EDEQALGIQLQKKIKELQARIEELEEEI-EAERASRAKAEKQRSDLS 1146
Score = 67.0 bits (162), Expect = 7e-11
Identities = 52/242 (21%), Positives = 113/242 (46%), Gaps = 1/242 (0%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
LE +R KN + E +LT+++ E + K+++E + +Q+ LEEA
Sbjct: 1501 LETLKRENKNLQQEISDLTEQIAEGGKRIHELEKIKKQVEQEKCELQAALEEAEASLEHE 1560
Query: 194 NEQAKKAVADSSRLFDEI-RQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXX 370
+ + + +++ E+ R+ E ++ID++K+ + +Q+ LD +E +
Sbjct: 1561 EGKILRIQLELNQVKSEVDRKIAEKDEEIDQLKRNHIRIVESMQSTLD-AEIRSRNDAIR 1619
Query: 371 XXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEK 550
E + E+E +L+ + E +N R + LK+ + D+ ++QE L++ +
Sbjct: 1620 LKKKMEGDLNEMEIQLNHANRMAAEALRNYRNTQGILKDTQIHLDDALRSQEDLKEQLAM 1679
Query: 551 LQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSSVST 730
++ + + ++EE + +QE+ D+ ER +Q L T+N S ++T
Sbjct: 1680 VERRANLLQAEIEELRATLEQTERSRKIAEQELLDASER-------VQLLHTQNTSLINT 1732
Query: 731 AR 736
+
Sbjct: 1733 KK 1734
Score = 66.2 bits (160), Expect = 1e-10
Identities = 49/229 (21%), Positives = 110/229 (48%), Gaps = 3/229 (1%)
Frame = +2
Query: 26 ERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQANEQA 205
++ ++N + E + E + + R L +L +++ EE+ ++ +
Sbjct: 1449 DKKQRNFDKILAEWKQKCEETHAELEASQKEARSLGTELFKIKNAYEESLDQLETLKREN 1508
Query: 206 KKAVADSSRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAMXXXXXXXXXX 385
K + S L ++I + + +++KIKKQ+E + ELQA L+E+E ++
Sbjct: 1509 KNLQQEISDLTEQIAEGGKRIHELEKIKKQVEQEKCELQAALEEAE-ASLEHEEGKILRI 1567
Query: 386 EQRVRELESELD---AEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQ 556
+ + +++SE+D AE+ ++ K R R ++ + D + +++ L +K++
Sbjct: 1568 QLELNQVKSEVDRKIAEKDEEIDQLK--RNHIRIVESMQSTLDAEIRSRNDAIRLKKKME 1625
Query: 557 GKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLR 703
G + + Q+ A +AA L YR Q ++D++ D A ++ + L+
Sbjct: 1626 GDLNEMEIQLNHANRMAAEALRNYRNTQGILKDTQIHLDDALRSQEDLK 1674
Score = 64.3 bits (155), Expect = 5e-10
Identities = 50/234 (21%), Positives = 107/234 (45%), Gaps = 7/234 (2%)
Frame = +2
Query: 17 ENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQAN 196
E+ E ++K +L D +EL T K+E + A ++ ++ + E +
Sbjct: 933 EDEEEINAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAGLD 992
Query: 197 E------QAKKAVADS-SRLFDEIRQEQEHAQQIDKIKKQLEAQNKELQAKLDESENNAM 355
E + KKA+ ++ + D+++ E++ + K K +LE Q +L+ L++ + M
Sbjct: 993 ETIAKLTKEKKALQEAHQQTLDDLQAEEDKVNTLTKAKIKLEQQVDDLEGSLEQEKKLRM 1052
Query: 356 XXXXXXXXXXEQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQ 535
E+ R+LE +L Q+ ++ + ++++ +LK+ + + E Q
Sbjct: 1053 DL--------ERAKRKLEGDLKLAQESIMDIENEKQQLDEKLKKKEFEISNLQSKIEDEQ 1104
Query: 536 DLVEKLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQK 697
L +LQ KIK + ++EE EE A K +++ D ++ + L++
Sbjct: 1105 ALGIQLQKKIKELQARIEELEEEIEAERASRAKAEKQRSDLSRELEEISERLEE 1158
Score = 45.4 bits (106), Expect = 2e-04
Identities = 49/221 (22%), Positives = 105/221 (47%), Gaps = 28/221 (12%)
Frame = +2
Query: 119 KRKLEADLAAMQSDLEEASNEARQA--NEQAKKAVADSSRLF--DEIRQEQEHAQQIDKI 286
K+K+E DL M+ L A+ A +A N + + + +++ D +R +++ +Q+ +
Sbjct: 1621 KKKMEGDLNEMEIQLNHANRMAAEALRNYRNTQGILKDTQIHLDDALRSQEDLKEQLAMV 1680
Query: 287 KKQ---LEAQNKELQAKLDESENNAMXXXXXXXXXXEQRVRELESELDAEQKRHVETQKN 457
+++ L+A+ +EL+A L+++E + +++ E E LDA ++ + +N
Sbjct: 1681 ERRANLLQAEIEELRATLEQTERS-------------RKIAEQEL-LDASERVQLLHTQN 1726
Query: 458 TRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTYKRQVEEAEEIA---------- 607
T I + K L+TD + ++QG+++ ++ AEE A
Sbjct: 1727 TSLINTKKK---LETD------------ISQMQGEMEDILQEARNAEEKAKKAITDAAMM 1771
Query: 608 ----------AVNLAKYRK-IQQEIEDSEERADQAEQALQK 697
+ +L + +K ++Q ++D + R D+AEQ K
Sbjct: 1772 AEELKKEQDTSAHLERMKKNMEQTVKDLQLRLDEAEQLALK 1812
Score = 41.2 bits (95), Expect = 0.004
Identities = 26/119 (21%), Positives = 58/119 (48%), Gaps = 7/119 (5%)
Frame = +2
Query: 386 EQRVRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKN----QERLQDLVE-- 547
++ ++++ EL + + E ++ + + ++ LQ + + +ER L++
Sbjct: 859 KEEFQKIKDELAKSEAKRKELEEKMVTLLKEKNDLQLQVQAEAEGLADAEERCDQLIKTK 918
Query: 548 -KLQGKIKTYKRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRSS 721
+L+ KIK + E+ EEI A AK RK++ E + ++ D E L K+ + ++
Sbjct: 919 IQLEAKIKEVTERAEDEEEINAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHAT 977
Score = 41.2 bits (95), Expect = 0.004
Identities = 58/288 (20%), Positives = 110/288 (38%), Gaps = 53/288 (18%)
Frame = +2
Query: 14 LENSERNRKNTESEKVELTDRLNELSVQSSSFMATKRKLEADLAAMQSDLEEASNEARQA 193
++N +R ++ E EK E+ +++L+ S + T K + +L M LE+ +E +
Sbjct: 1213 IDNLQRVKQKLEKEKSEMKMEIDDLA----SNVETVSKAKGNLEKMCRTLEDQLSELKSK 1268
Query: 194 NEQAKKAVADS--------------SRLFDE--------IRQEQEHAQQIDKIKKQLEAQ 307
E+ ++ + D SR DE R +Q QQI+++K+QLE +
Sbjct: 1269 EEEQQRLINDLTAQRGRLQTESGEFSRQLDEKEALVSQLSRGKQAFTQQIEELKRQLEEE 1328
Query: 308 NK-------------------------------ELQAKLDESENNAMXXXXXXXXXXEQR 394
K ELQ L ++ QR
Sbjct: 1329 IKAKNALAHALQSSRHDCDLLREQYEEEQESKAELQRALSKANTEVAQWRTKYETDAIQR 1388
Query: 395 VRELESELDAEQKRHVETQKNTRKIERRLKEIGLQTDEDKKNQERLQDLVEKLQGKIKTY 574
ELE +A++K + + E ++ + + +K ++RLQ+ VE L
Sbjct: 1389 TEELE---EAKKK----LAQRLQAAEEHVEAVNAKCASLEKTKQRLQNEVEDLM------ 1435
Query: 575 KRQVEEAEEIAAVNLAKYRKIQQEIEDSEERADQAEQALQKLRTKNRS 718
VE A K R + + + +++ ++ L+ + + RS
Sbjct: 1436 -LDVERTNAACAALDKKQRNFDKILAEWKQKCEETHAELEASQKEARS 1482
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 86,284,299
Number of Sequences: 369166
Number of extensions: 1497959
Number of successful extensions: 11544
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8273
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10362
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 9895030780
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail