DrC_00042
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
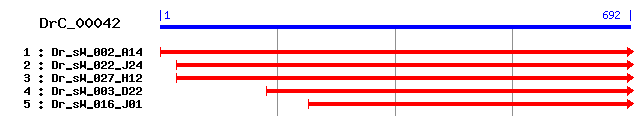
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00042 (692 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P24534|EF1B_HUMAN Elongation factor 1-beta (EF-1-beta) 253 3e-67 sp|P34826|EF1B_RABIT Elongation factor 1-beta (EF-1-beta) 253 3e-67 sp|Q5E983|EF1B_BOVIN Elongation factor 1-beta (EF-1-beta) 249 5e-66 sp|O70251|EF1B_MOUSE Elongation factor 1-beta (EF-1-beta) 248 1e-65 sp|Q9YGQ1|EF1B_CHICK Elongation factor 1-beta (EF-1-beta) 247 2e-65 sp|Q6DET9|EF1B_XENTR Elongation factor 1-beta (EF-1-beta) 239 4e-63 sp|P30151|EF1B_XENLA Elongation factor 1-beta (EF-1-beta) (... 236 4e-62 sp|O96827|EF1B_DROME Probable elongation factor 1-beta (EF-... 230 2e-60 sp|P12262|EF1B_ARTSA Elongation factor 1-beta (EF-1-beta) 228 9e-60 sp|P29522|EF1B2_BOMMO Elongation factor 1-beta' 222 6e-58
>sp|P24534|EF1B_HUMAN Elongation factor 1-beta (EF-1-beta) Length = 225 Score = 253 bits (647), Expect = 3e-67 Identities = 136/225 (60%), Positives = 156/225 (69%), Gaps = 17/225 (7%) Frame = +2 Query: 20 MEFGNLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFHGCESIKDC-YCHALRWFNHIK 196 M FG+LK G LNDYL DKSYI GY PS AD+ VF S CHALRW+NHIK Sbjct: 1 MGFGDLKSPAGLQVLNDYLADKSYIEGYVPSQADVAVFEAVSSPPPADLCHALRWYNHIK 60 Query: 197 SFSEAE------KKAFRHVEPV---------AQXXXXXXXVDLFGSDDEED-EEAARIKQ 328 S+ + + KKA P A +DLFGSDDEE+ EEA R+++ Sbjct: 61 SYEKEKASLPGVKKALGKYGPADVEDTTGSGATDSKDDDDIDLFGSDDEEESEEAKRLRE 120 Query: 329 ERLDAYAAKKSTKPALVAKSSITLDVKPWDDETDMVELEKCVRSITADGLLWGASKFAPV 508 ERL Y +KK+ KPALVAKSSI LDVKPWDDETDM +LE+CVRSI ADGL+WG+SK PV Sbjct: 121 ERLAQYESKKAKKPALVAKSSILLDVKPWDDETDMAKLEECVRSIQADGLVWGSSKLVPV 180 Query: 509 GYGIRKLVITCVVEDDKVGTDFLEEEITKFEDYVQSMDVVSFNKI 643 GYGI+KL I CVVEDDKVGTD LEE+IT FEDYVQSMDV +FNKI Sbjct: 181 GYGIKKLQIQCVVEDDKVGTDMLEEQITAFEDYVQSMDVAAFNKI 225
>sp|P34826|EF1B_RABIT Elongation factor 1-beta (EF-1-beta) Length = 225 Score = 253 bits (646), Expect = 3e-67 Identities = 136/225 (60%), Positives = 156/225 (69%), Gaps = 17/225 (7%) Frame = +2 Query: 20 MEFGNLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFHGCESIKDC-YCHALRWFNHIK 196 M FG+LK G LNDYL DKSYI GY PS AD+ VF CHALRW+NHIK Sbjct: 1 MGFGDLKSPAGLQVLNDYLADKSYIEGYVPSQADVAVFEAVSGPPPADLCHALRWYNHIK 60 Query: 197 SFSEAE------KKAFRHVEPV---------AQXXXXXXXVDLFGSDDEED-EEAARIKQ 328 S+ + + KKA P A +DLFGSDDEE+ EEA R+++ Sbjct: 61 SYEKEKASLPGIKKALGTYGPADVEDTTGSGATDSKDDDDIDLFGSDDEEESEEAKRLRE 120 Query: 329 ERLDAYAAKKSTKPALVAKSSITLDVKPWDDETDMVELEKCVRSITADGLLWGASKFAPV 508 ERL Y +KK+ KPALVAKSSI LDVKPWDDETDMV+LE+CVRSI ADGL+WG+SK PV Sbjct: 121 ERLAQYESKKAKKPALVAKSSILLDVKPWDDETDMVKLEECVRSIQADGLVWGSSKLVPV 180 Query: 509 GYGIRKLVITCVVEDDKVGTDFLEEEITKFEDYVQSMDVVSFNKI 643 GYGI+KL I CVVEDDKVGTD LEE+IT FEDYVQSMDV +FNKI Sbjct: 181 GYGIKKLQIQCVVEDDKVGTDMLEEQITAFEDYVQSMDVAAFNKI 225
>sp|Q5E983|EF1B_BOVIN Elongation factor 1-beta (EF-1-beta) Length = 225 Score = 249 bits (636), Expect = 5e-66 Identities = 133/225 (59%), Positives = 155/225 (68%), Gaps = 17/225 (7%) Frame = +2 Query: 20 MEFGNLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFHGCESIKDC-YCHALRWFNHIK 196 M FG+LK G LNDYL DKSYI GY PS AD+ VF CHALRW+NHIK Sbjct: 1 MGFGDLKSPAGLQVLNDYLADKSYIEGYVPSQADVAVFEAVSGPPPADLCHALRWYNHIK 60 Query: 197 SFSEAE------KKAFRHVEPV---------AQXXXXXXXVDLFGSDDEED-EEAARIKQ 328 S+ + + KKA P A +DLFGSDDEE+ EEA R+++ Sbjct: 61 SYEKEKASLPGVKKALGKYGPANVEDTTESGATDSKDDDDIDLFGSDDEEESEEAKRLRE 120 Query: 329 ERLDAYAAKKSTKPALVAKSSITLDVKPWDDETDMVELEKCVRSITADGLLWGASKFAPV 508 ERL Y +KK+ KPALVAKSSI LDVKPWDDETDM +LE+CVRSI ADGL+WG+SK PV Sbjct: 121 ERLAQYESKKAKKPALVAKSSILLDVKPWDDETDMAKLEECVRSIQADGLVWGSSKLVPV 180 Query: 509 GYGIRKLVITCVVEDDKVGTDFLEEEITKFEDYVQSMDVVSFNKI 643 GYGI+KL I CVVEDDKVGTD LEE+IT F++YVQSMDV +FNKI Sbjct: 181 GYGIKKLQIQCVVEDDKVGTDMLEEQITAFDEYVQSMDVAAFNKI 225
>sp|O70251|EF1B_MOUSE Elongation factor 1-beta (EF-1-beta) Length = 225 Score = 248 bits (633), Expect = 1e-65 Identities = 132/225 (58%), Positives = 155/225 (68%), Gaps = 17/225 (7%) Frame = +2 Query: 20 MEFGNLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFHGCESIKDC-YCHALRWFNHIK 196 M FG+LK G LNDYL DKSYI GY PS AD+ VF CHALRW+NHIK Sbjct: 1 MGFGDLKTPAGLQVLNDYLADKSYIEGYVPSQADVAVFEAVSGPPPADLCHALRWYNHIK 60 Query: 197 SFSEAE------KKAFRHVEPV---------AQXXXXXXXVDLFGSDDEED-EEAARIKQ 328 S+ + + KK+ P A +DLFGSDDEE+ EEA ++++ Sbjct: 61 SYEKEKASLPGVKKSLGKYGPSSVEDTTGSGAADAKDDDDIDLFGSDDEEESEEAKKLRE 120 Query: 329 ERLDAYAAKKSTKPALVAKSSITLDVKPWDDETDMVELEKCVRSITADGLLWGASKFAPV 508 ERL Y +KK+ KPA+VAKSSI LDVKPWDDETDM +LE+CVRSI ADGL+WG+SK PV Sbjct: 121 ERLAQYESKKAKKPAVVAKSSILLDVKPWDDETDMTKLEECVRSIQADGLVWGSSKLVPV 180 Query: 509 GYGIRKLVITCVVEDDKVGTDFLEEEITKFEDYVQSMDVVSFNKI 643 GYGI+KL I CVVEDDKVGTD LEE+IT FEDYVQSMDV +FNKI Sbjct: 181 GYGIKKLQIQCVVEDDKVGTDMLEEQITAFEDYVQSMDVAAFNKI 225
>sp|Q9YGQ1|EF1B_CHICK Elongation factor 1-beta (EF-1-beta) Length = 225 Score = 247 bits (631), Expect = 2e-65 Identities = 135/226 (59%), Positives = 157/226 (69%), Gaps = 18/226 (7%) Frame = +2 Query: 20 MEFGNLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFH--GCESIKDCYCHALRWFNHI 193 M FG+LK G LND+L DKSYI GY PS AD+ VF G D + HALRW+NHI Sbjct: 1 MGFGDLKSAAGLRVLNDFLADKSYIEGYVPSQADIAVFEAVGAPPPADLF-HALRWYNHI 59 Query: 194 KSFSEAE------KKAFRHVEPV---------AQXXXXXXXVDLFGSDDEED-EEAARIK 325 KS+ + + KKA P A +DLFGSDDEE+ EEA R++ Sbjct: 60 KSYEKEKASLPGVKKALGKYGPADVEDTTGSGATDSKDDDDIDLFGSDDEEESEEAKRLR 119 Query: 326 QERLDAYAAKKSTKPALVAKSSITLDVKPWDDETDMVELEKCVRSITADGLLWGASKFAP 505 +ERL Y +KK+ KPALVAKSSI LDVKPWDDETDM +LE+CVRSI ADGL+WG+SK P Sbjct: 120 EERLAQYESKKAKKPALVAKSSILLDVKPWDDETDMAKLEECVRSIQADGLVWGSSKLVP 179 Query: 506 VGYGIRKLVITCVVEDDKVGTDFLEEEITKFEDYVQSMDVVSFNKI 643 VGYGI+KL I CVVEDDKVGTD LEE+IT FEDYVQSMDV +FNKI Sbjct: 180 VGYGIKKLQIQCVVEDDKVGTDMLEEQITAFEDYVQSMDVAAFNKI 225
>sp|Q6DET9|EF1B_XENTR Elongation factor 1-beta (EF-1-beta) Length = 228 Score = 239 bits (611), Expect = 4e-63 Identities = 129/228 (56%), Positives = 153/228 (67%), Gaps = 20/228 (8%) Frame = +2 Query: 20 MEFGNLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFHGCESIKDC-YCHALRWFNHIK 196 M FG+LK G LN++L DKSYI GY PS AD+ VF HALRW+NHIK Sbjct: 1 MGFGDLKSPAGLKVLNEFLADKSYIEGYVPSQADVAVFDALSGAPPADLFHALRWYNHIK 60 Query: 197 SFSEAE------KKAFRHVEPV------------AQXXXXXXXVDLFGSDDEED-EEAAR 319 S+ + + KK + PV + +DLFGSDDEE+ EE+ R Sbjct: 61 SYEKQKSSLPGVKKPLGNYGPVNIEDTTGSTAKDTKEEDDDDDIDLFGSDDEEENEESKR 120 Query: 320 IKQERLDAYAAKKSTKPALVAKSSITLDVKPWDDETDMVELEKCVRSITADGLLWGASKF 499 +++ERL Y AKKS KPAL+AKSSI LDVKPWDDETDM +LE+CVRSI +GL+WGASK Sbjct: 121 VREERLAQYEAKKSKKPALIAKSSILLDVKPWDDETDMAKLEECVRSIQMEGLVWGASKL 180 Query: 500 APVGYGIRKLVITCVVEDDKVGTDFLEEEITKFEDYVQSMDVVSFNKI 643 PVGYGI+KL I CVVEDDKVGTD LEE IT FED+VQSMDV +FNKI Sbjct: 181 VPVGYGIKKLQIQCVVEDDKVGTDVLEENITAFEDFVQSMDVAAFNKI 228
>sp|P30151|EF1B_XENLA Elongation factor 1-beta (EF-1-beta) (p30) Length = 227 Score = 236 bits (602), Expect = 4e-62 Identities = 128/227 (56%), Positives = 152/227 (66%), Gaps = 19/227 (8%) Frame = +2 Query: 20 MEFGNLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFHGCESIKDC-YCHALRWFNHIK 196 M FG+LK G L ++L DKSYI GY PS AD+ VF + HALRW+NHIK Sbjct: 1 MGFGDLKSPAGLKVLKEFLADKSYIEGYVPSQADVAVFDALSAAPPADLFHALRWYNHIK 60 Query: 197 SFSEAE------KKAFRHVEPV-----------AQXXXXXXXVDLFGSDDEED-EEAARI 322 S+ + + KKA + PV +DLFGSDDEE+ E+A R+ Sbjct: 61 SYEKQKSSLPGVKKALGNYGPVNIEDTTGSAAKETKEEDDDDIDLFGSDDEEESEDAKRV 120 Query: 323 KQERLDAYAAKKSTKPALVAKSSITLDVKPWDDETDMVELEKCVRSITADGLLWGASKFA 502 + ERL Y AKKS KP L+AKSSI LDVKPWDDETDM +LE+C+RSI DGLLWG+SK Sbjct: 121 RDERLAQYEAKKSKKPTLIAKSSILLDVKPWDDETDMGKLEECLRSIQMDGLLWGSSKLV 180 Query: 503 PVGYGIRKLVITCVVEDDKVGTDFLEEEITKFEDYVQSMDVVSFNKI 643 PVGYGI+KL I CVVEDDKVGTD LEE+IT FED+VQSMDV +FNKI Sbjct: 181 PVGYGIKKLQIQCVVEDDKVGTDVLEEKITAFEDFVQSMDVAAFNKI 227
>sp|O96827|EF1B_DROME Probable elongation factor 1-beta (EF-1-beta) Length = 222 Score = 230 bits (587), Expect = 2e-60 Identities = 124/223 (55%), Positives = 147/223 (65%), Gaps = 15/223 (6%) Frame = +2 Query: 20 MEFGNLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFHGCESIKDC-YCHALRWFNHIK 196 M FG++ G +LN +L D SYI+GYTPS ADL VF + RW+ HI Sbjct: 1 MAFGDVTTPQGLKELNAFLADNSYISGYTPSKADLSVFDALGKAPSADNVNVARWYRHIA 60 Query: 197 SFSEAEKKAFRHVEPVAQXXXXXXXV--------------DLFGSDDEEDEEAARIKQER 334 SF AE+ A+ P+ Q V DLFGSDDEEDEEA RIKQER Sbjct: 61 SFEAAERAAWSGT-PLPQLAGGKPTVAAAAKPAADDDDDVDLFGSDDEEDEEAERIKQER 119 Query: 335 LDAYAAKKSTKPALVAKSSITLDVKPWDDETDMVELEKCVRSITADGLLWGASKFAPVGY 514 + AYAAKKS KPAL+AKSS+ LDVKPWDDETDM E+E VR+I DGLLWGASK PVGY Sbjct: 120 VAAYAAKKSKKPALIAKSSVLLDVKPWDDETDMKEMENNVRTIEMDGLLWGASKLVPVGY 179 Query: 515 GIRKLVITCVVEDDKVGTDFLEEEITKFEDYVQSMDVVSFNKI 643 GI KL I CV+EDDKV D L+E+I +FED+VQS+D+ +FNKI Sbjct: 180 GINKLQIMCVIEDDKVSIDLLQEKIEEFEDFVQSVDIAAFNKI 222
>sp|P12262|EF1B_ARTSA Elongation factor 1-beta (EF-1-beta) Length = 207 Score = 228 bits (582), Expect = 9e-60 Identities = 114/205 (55%), Positives = 149/205 (72%), Gaps = 1/205 (0%) Frame = +2 Query: 32 NLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFHGCESI-KDCYCHALRWFNHIKSFSE 208 +LK + GQ +LN+ L +KSY+ GY PS D+ F+ D + + LRW+ HI SFS+ Sbjct: 5 DLKAEKGQEQLNELLANKSYLQGYEPSQEDVAAFNQLNKAPSDKFPYLLRWYKHISSFSD 64 Query: 209 AEKKAFRHVEPVAQXXXXXXXVDLFGSDDEEDEEAARIKQERLDAYAAKKSTKPALVAKS 388 AEKK F + P + VDLFGSD EEDEEA +IK ER+ AY+ KKS KPA+VAKS Sbjct: 65 AEKKGFPGI-PTSASKEEDDDVDLFGSD-EEDEEAEKIKAERMKAYSDKKSKKPAIVAKS 122 Query: 389 SITLDVKPWDDETDMVELEKCVRSITADGLLWGASKFAPVGYGIRKLVITCVVEDDKVGT 568 S+ LD+KPWDDETDM E+EK VRS+ DGL+WGA+K P+ YGI+KL I CVVEDDKV Sbjct: 123 SVILDIKPWDDETDMAEMEKLVRSVQMDGLVWGAAKLIPLAYGIKKLSIMCVVEDDKVSI 182 Query: 569 DFLEEEITKFEDYVQSMDVVSFNKI 643 D L+E+I++FED+VQS+D+ +FNK+ Sbjct: 183 DELQEKISEFEDFVQSVDIAAFNKV 207
>sp|P29522|EF1B2_BOMMO Elongation factor 1-beta' Length = 222 Score = 222 bits (566), Expect = 6e-58 Identities = 120/222 (54%), Positives = 148/222 (66%), Gaps = 14/222 (6%) Frame = +2 Query: 20 MEFGNLKIDIGQSKLNDYLFDKSYINGYTPSSADLEVFHGCESIKDCYC-HALRWFNHIK 196 M G++K G + LN YL +KSY++GYTPS AD++VF H LRW+N I Sbjct: 1 MAVGDVKTAQGLNDLNQYLAEKSYVSGYTPSQADVQVFEQVGKAPAANLPHVLRWYNQIA 60 Query: 197 SFSEAEKKAFRH--------VEPVAQXXXXXXX----VDLFGS-DDEEDEEAARIKQERL 337 S++ AE+K + +P A VDLFGS D+EED EA RI++ERL Sbjct: 61 SYTSAERKTWSQGTSPLTAGAKPTAPAPAAKDDDDDDVDLFGSGDEEEDAEAERIREERL 120 Query: 338 DAYAAKKSTKPALVAKSSITLDVKPWDDETDMVELEKCVRSITADGLLWGASKFAPVGYG 517 AYA KKS KPAL+AKSSI LDVKPWDDETDM E+E VR+I +GLLWGASK PVGYG Sbjct: 121 KAYADKKSKKPALIAKSSILLDVKPWDDETDMKEMENQVRTIEMEGLLWGASKLVPVGYG 180 Query: 518 IRKLVITCVVEDDKVGTDFLEEEITKFEDYVQSMDVVSFNKI 643 I KL I CV+EDDKV D L E+I +FED+VQS+D+ +FNKI Sbjct: 181 INKLQIMCVIEDDKVSVDLLTEKIQEFEDFVQSVDIAAFNKI 222
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,015,725
Number of Sequences: 369166
Number of extensions: 1252580
Number of successful extensions: 3519
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3334
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3445
length of database: 68,354,980
effective HSP length: 107
effective length of database: 48,588,335
effective search space used: 5976365205
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail