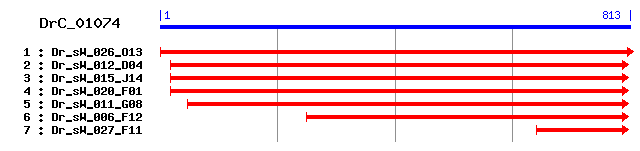
DrC_01074
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01074 (813 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O00463|TRAF5_HUMAN TNF receptor-associated factor 5 (RIN... 59 2e-08 sp|P70196|TRAF6_MOUSE TNF receptor-associated factor 6 57 5e-08 sp|P70191|TRAF5_MOUSE TNF receptor-associated factor 5 57 6e-08 sp|Q9Y4K3|TRAF6_HUMAN TNF receptor-associated factor 6 (Int... 56 1e-07 sp|P68907|PZRN3_RAT PDZ domain-containing RING finger prote... 54 5e-07 sp|Q69ZS0|PZRN3_MOUSE PDZ domain-containing RING finger pro... 52 2e-06 sp|Q9UPQ7|PZRN3_HUMAN PDZ domain-containing RING finger pro... 49 1e-05 sp|Q13114|TRAF3_HUMAN TNF receptor-associated factor 3 (CD4... 47 6e-05 sp|Q60803|TRAF3_MOUSE TNF receptor-associated factor 3 (CD4... 47 6e-05 sp|Q9BUZ4|TRAF4_HUMAN TNF receptor-associated factor 4 (Cys... 46 1e-04
>sp|O00463|TRAF5_HUMAN TNF receptor-associated factor 5 (RING finger protein 84) Length = 557 Score = 58.5 bits (140), Expect = 2e-08 Identities = 38/140 (27%), Positives = 67/140 (47%), Gaps = 6/140 (4%) Frame = +2 Query: 221 SACSGNTEECKLENISQTVINHRLNEDISNLTVKCPEEK--CSFECRLI----DMDEHLK 382 ++C E+C VIN + +E+ NL CPE C C I ++DEHL Sbjct: 159 ASCQFRKEKCLYCKKDVVVINLQNHEE--NL---CPEYPVFCPNNCAKIILKTEVDEHLA 213 Query: 383 VCDNKTEKCPFFGYGCTHKIKSKNMNSHMLREAQDHNNMLIDLLGNLKNEVIDLREKEKN 562 VC + CPF YGC K +N+ H ++H ++++ L+ ++ DL + + Sbjct: 214 VCPEAEQDCPFKHYGCAVTDKRRNLQQHEHSALREHMRLVLEKNVQLEEQISDLHKSLEQ 273 Query: 563 NHDEIEKLKEKGRVQDEKLK 622 +I++L E + +++ K Sbjct: 274 KESKIQQLAETIKKLEKEFK 293
Score = 40.0 bits (92), Expect = 0.008
Identities = 34/162 (20%), Positives = 65/162 (40%), Gaps = 2/162 (1%)
Frame = +2
Query: 83 KFSVIGEITNDMKCTLCGNIAVNAQQALCGCNFCNKCITNFIN-NGGSACSGNTEECKLE 259
++ + + KC C ++ N Q CG FC CI + N C + E K +
Sbjct: 32 EYQFVERLEERYKCAFCHSVLHNPHQTGCGHRFCQHCILSLRELNTVPICPVDKEVIKSQ 91
Query: 260 NISQTVINHRLNEDISNLTVKCPEEK-CSFECRLIDMDEHLKVCDNKTEKCPFFGYGCTH 436
+ + ++ ++ NL V C C+ + L +HL+ C + +C C
Sbjct: 92 EVFK---DNCCKREVLNLYVYCSNAPGCNAKVILGRYQDHLQQCLFQPVQCS--NEKCRE 146
Query: 437 KIKSKNMNSHMLREAQDHNNMLIDLLGNLKNEVIDLREKEKN 562
+ K++ H+ Q + ++ VI+L+ E+N
Sbjct: 147 PVLRKDLKEHLSASCQFRKEKCLYCKKDV--VVINLQNHEEN 186
>sp|P70196|TRAF6_MOUSE TNF receptor-associated factor 6 Length = 530 Score = 57.4 bits (137), Expect = 5e-08 Identities = 39/152 (25%), Positives = 64/152 (42%) Frame = +2 Query: 119 KCTLCGNIAVNAQQALCGCNFCNKCITNFINNGGSACSGNTEECKLENISQTVINHRLNE 298 +C +C A Q CG FC CI I + G C + E LEN Q ++ Sbjct: 69 ECPICLMALREAVQTPCGHRFCKACIIKSIRDAGHKCPVD-NEILLEN--QLFPDNFAKR 125 Query: 299 DISNLTVKCPEEKCSFECRLIDMDEHLKVCDNKTEKCPFFGYGCTHKIKSKNMNSHMLRE 478 +I +LTVKCP + C + L +++H C+ CP C + +N+H++ + Sbjct: 126 EILSLTVKCPNKGCLQKMELRHLEDHQVHCEFALVNCP----QCQRPFQKCQVNTHIIED 181 Query: 479 AQDHNNMLIDLLGNLKNEVIDLREKEKNNHDE 574 + N + + +EK HD+ Sbjct: 182 CPRRQVSCV-------NCAVSMAYEEKEIHDQ 206
>sp|P70191|TRAF5_MOUSE TNF receptor-associated factor 5 Length = 558 Score = 57.0 bits (136), Expect = 6e-08 Identities = 35/125 (28%), Positives = 59/125 (47%) Frame = +2 Query: 248 CKLENISQTVINHRLNEDISNLTVKCPEEKCSFECRLIDMDEHLKVCDNKTEKCPFFGYG 427 CK + + + +H N V CP +C ++EHL VC + CPF YG Sbjct: 171 CKRDIVVTNLQDHEENS-CPAYPVSCPN-RCVQTIPRARVNEHLTVCPEAEQDCPFKHYG 228 Query: 428 CTHKIKSKNMNSHMLREAQDHNNMLIDLLGNLKNEVIDLREKEKNNHDEIEKLKEKGRVQ 607 CT K K N+ H QDH ++++ L+ + DL + + +I++L E + Sbjct: 229 CTVKGKRGNLLEHERAALQDHMLLVLEKNYQLEQRISDLYQSLEQKESKIQQLAETVKKF 288 Query: 608 DEKLK 622 +++LK Sbjct: 289 EKELK 293
Score = 45.8 bits (107), Expect = 1e-04
Identities = 30/133 (22%), Positives = 55/133 (41%), Gaps = 2/133 (1%)
Frame = +2
Query: 77 DHKFSVIGEITNDMKCTLCGNIAVNAQQALCGCNFCNKCITNFIN-NGGSACSGNTEECK 253
D ++ + ++ KC C ++ N Q CG FC +CI + N C + E K
Sbjct: 30 DTEYQFVEQLEERYKCAFCHSVLHNPHQTGCGHRFCQQCIRSLRELNSVPICPVDKEVIK 89
Query: 254 LENISQTVINHRLNEDISNLTVKCPEEK-CSFECRLIDMDEHLKVCDNKTEKCPFFGYGC 430
+ + + ++ ++ NL V C C+ L +HL+ C + CP C
Sbjct: 90 PQEVFK---DNCCKREVLNLHVYCKNAPGCNARIILGRFQDHLQHCSFQAVPCP--NESC 144
Query: 431 THKIKSKNMNSHM 469
+ K++ H+
Sbjct: 145 REAMLRKDVKEHL 157
>sp|Q9Y4K3|TRAF6_HUMAN TNF receptor-associated factor 6 (Interleukin 1 signal transducer) (RING finger protein 85) Length = 522 Score = 56.2 bits (134), Expect = 1e-07 Identities = 35/120 (29%), Positives = 54/120 (45%) Frame = +2 Query: 119 KCTLCGNIAVNAQQALCGCNFCNKCITNFINNGGSACSGNTEECKLENISQTVINHRLNE 298 +C +C A Q CG FC CI I + G C + E LEN Q ++ Sbjct: 69 ECPICLMALREAVQTPCGHRFCKACIIKSIRDAGHKCPVD-NEILLEN--QLFPDNFAKR 125 Query: 299 DISNLTVKCPEEKCSFECRLIDMDEHLKVCDNKTEKCPFFGYGCTHKIKSKNMNSHMLRE 478 +I +L VKCP E C + L +++H C+ CP C + ++N H+L++ Sbjct: 126 EILSLMVKCPNEGCLHKMELRHLEDHQAHCEFALMDCP----QCQRPFQKFHINIHILKD 181
>sp|P68907|PZRN3_RAT PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein) Length = 1062 Score = 53.9 bits (128), Expect = 5e-07 Identities = 40/134 (29%), Positives = 61/134 (45%), Gaps = 4/134 (2%) Frame = +2 Query: 98 GEITNDMKCTLCGNIAVNAQQALCGCNFCNKCITNFINNGGSACSGNTEECKLENISQTV 277 GE+ D+KC LC + + CG FC C+ ++ GS S C+ +S Sbjct: 10 GEVDPDLKCALCHKVLEDPLTTPCGHVFCAGCVLPWVVQEGSCPS----RCR-GRLSAKE 64 Query: 278 INH--RLNEDISNLTVKCPE--EKCSFECRLIDMDEHLKVCDNKTEKCPFFGYGCTHKIK 445 +NH L I L +KC C +L D+ EHL+ CD +C GC + Sbjct: 65 LNHVLPLKRLILKLDIKCAHAARGCGRVVKLQDLPEHLERCDFAPARCRH--AGCGQLLL 122 Query: 446 SKNMNSHMLREAQD 487 +++ +HM R+A D Sbjct: 123 RRDVEAHM-RDACD 135
>sp|Q69ZS0|PZRN3_MOUSE PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein) Length = 1063 Score = 52.0 bits (123), Expect = 2e-06 Identities = 39/137 (28%), Positives = 60/137 (43%), Gaps = 7/137 (5%) Frame = +2 Query: 98 GEITNDMKCTLCGNIAVNAQQALCGCNFCNKCITNFINNGGSA---CSGNTEECKLENIS 268 G++ D+KC LC + + CG FC C+ ++ GS C G +S Sbjct: 10 GDVDPDLKCALCHKVLEDPLTTPCGHVFCAGCVLPWVVQEGSCPARCRG--------RLS 61 Query: 269 QTVINH--RLNEDISNLTVKCPE--EKCSFECRLIDMDEHLKVCDNKTEKCPFFGYGCTH 436 +NH L I L +KC C +L D+ EHL+ CD +C GC Sbjct: 62 AKELNHVLPLKRLILKLDIKCAHAARGCGRVVKLQDLPEHLERCDFAPARCRH--AGCGQ 119 Query: 437 KIKSKNMNSHMLREAQD 487 + +++ +HM R+A D Sbjct: 120 LLLRRDVEAHM-RDACD 135
>sp|Q9UPQ7|PZRN3_HUMAN PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein) Length = 1066 Score = 49.3 bits (116), Expect = 1e-05 Identities = 38/137 (27%), Positives = 59/137 (43%), Gaps = 7/137 (5%) Frame = +2 Query: 98 GEITNDMKCTLCGNIAVNAQQALCGCNFCNKCITNFINNGGSA---CSGNTEECKLENIS 268 G++ D+KC LC + + CG FC C+ ++ GS C G +S Sbjct: 10 GDVDPDLKCALCHKVLEDPLTTPCGHVFCAGCVLPWVVQEGSCPARCRG--------RLS 61 Query: 269 QTVINH--RLNEDISNLTVKC--PEEKCSFECRLIDMDEHLKVCDNKTEKCPFFGYGCTH 436 +NH L I L +KC C +L + EHL+ CD +C GC Sbjct: 62 AKELNHVLPLKRLILKLDIKCAYATRGCGRVVKLQQLPEHLERCDFAPARCRH--AGCGQ 119 Query: 437 KIKSKNMNSHMLREAQD 487 + +++ +HM R+A D Sbjct: 120 VLLRRDVEAHM-RDACD 135
>sp|Q13114|TRAF3_HUMAN TNF receptor-associated factor 3 (CD40 receptor associated factor 1) (CRAF1) (CD40 binding protein) (CD40BP) (LMP1 associated protein) (LAP1) (CAP-1) Length = 568 Score = 47.0 bits (110), Expect = 6e-05 Identities = 33/134 (24%), Positives = 59/134 (44%), Gaps = 4/134 (2%) Frame = +2 Query: 248 CKLENISQTVINHRLNEDISNLTVKCPEEKCSFECRL-IDMDEHLKVCDNKTEKCPFFGY 424 CK + + + + D + V CP KCS + L ++ HL C N C F Y Sbjct: 180 CKSQ-VPMIALQKHEDTDCPCVVVSCPH-KCSVQTLLRSELSAHLSECVNAPSTCSFKRY 237 Query: 425 GCTHKIKSKNMNSHMLREAQDHNNMLIDLLGNLKNEVIDLREKEKNNHDEIEKLKEK--- 595 GC + ++ + +H A H N+L + +L+ +V L+ + + I+ L + Sbjct: 238 GCVFQGTNQQIKAHEASSAVQHVNLLKEWSNSLEKKVSLLQNESVEKNKSIQSLHNQICS 297 Query: 596 GRVQDEKLKAMYEN 637 ++ E+ K M N Sbjct: 298 FEIEIERQKEMLRN 311
Score = 41.6 bits (96), Expect = 0.003
Identities = 28/132 (21%), Positives = 55/132 (41%), Gaps = 2/132 (1%)
Frame = +2
Query: 80 HKFSVIGEITNDMKCTLCGNIAVNAQQALCGCNFCNKCITNFINNGGSACSGNTEECKLE 259
+K + + + KC C + + +Q CG FC C+ +++ C+ E +
Sbjct: 39 YKEKFVKTVEDKYKCEKCHLVLCSPKQTECGHRFCESCMAALLSSSSPKCTACQESIVKD 98
Query: 260 NISQTVINHRLNEDISNLTVKCPEEK--CSFECRLIDMDEHLKVCDNKTEKCPFFGYGCT 433
+ + ++ +I L + C E C+ + L + HLK D E+ P C
Sbjct: 99 KVFK---DNCCKREILALQIYCRNESRGCAEQLTLGHLLVHLK-NDCHFEELPCVRPDCK 154
Query: 434 HKIKSKNMNSHM 469
K+ K++ H+
Sbjct: 155 EKVLRKDLRDHV 166
>sp|Q60803|TRAF3_MOUSE TNF receptor-associated factor 3 (CD40 receptor associated factor 1) (CRAF1) (TRAFAMN) Length = 567 Score = 47.0 bits (110), Expect = 6e-05 Identities = 36/143 (25%), Positives = 59/143 (41%), Gaps = 4/143 (2%) Frame = +2 Query: 221 SACSGNTEECKLENISQTVINHRLNEDISNLTVKCPEEKCSFECRL-IDMDEHLKVCDNK 397 S C KL+ T D + V CP KCS + L ++ HL C N Sbjct: 177 SHCKSQVPMIKLQKHEDT--------DCPCVVVSCPH-KCSVQTLLRSELSAHLSECVNA 227 Query: 398 TEKCPFFGYGCTHKIKSKNMNSHMLREAQDHNNMLIDLLGNLKNEVIDLREKEKNNHDEI 577 C F YGC + ++ + +H A H N+L + +L+ +V L+ + + I Sbjct: 228 PSTCSFKRYGCVFQGTNQQIKAHEASSAVQHVNLLKEWSNSLEKKVSLLQNESVEKNKSI 287 Query: 578 EKLKEK---GRVQDEKLKAMYEN 637 + L + ++ E+ K M N Sbjct: 288 QSLHNQICSFEIEIERQKEMLRN 310
Score = 42.7 bits (99), Expect = 0.001
Identities = 29/132 (21%), Positives = 56/132 (42%), Gaps = 2/132 (1%)
Frame = +2
Query: 80 HKFSVIGEITNDMKCTLCGNIAVNAQQALCGCNFCNKCITNFINNGGSACSGNTEECKLE 259
+K + + + KC C + N +Q CG FC C+ +++ C+ E +
Sbjct: 38 YKEKFVKTVEDKYKCEKCRLVLCNPKQTECGHRFCESCMAALLSSSSPKCTACQESIIKD 97
Query: 260 NISQTVINHRLNEDISNLTVKCPEE--KCSFECRLIDMDEHLKVCDNKTEKCPFFGYGCT 433
+ + ++ +I L V C E C+ + L + HLK + + E+ P C
Sbjct: 98 KVFK---DNCCKREILALQVYCRNEGRGCAEQLTLGHLLVHLK-NECQFEELPCLRADCK 153
Query: 434 HKIKSKNMNSHM 469
K+ K++ H+
Sbjct: 154 EKVLRKDLRDHV 165
>sp|Q9BUZ4|TRAF4_HUMAN TNF receptor-associated factor 4 (Cysteine-rich domain associated with RING and Traf domains protein 1) (Malignant 62) (RING finger protein 83) Length = 470 Score = 45.8 bits (107), Expect = 1e-04 Identities = 40/162 (24%), Positives = 64/162 (39%), Gaps = 16/162 (9%) Frame = +2 Query: 116 MKCTLCG----NIAVNAQQALCGCN--FC-NKCITNFIN-----NGGSACSGNTEECKL- 256 +KC CG A + + +C +C NKC + + S C T+ C Sbjct: 139 LKCEFCGCDFSGEAYESHEGMCPQESVYCENKCGARMMRRLLAQHATSECPKRTQPCTYC 198 Query: 257 --ENISQTVINHRLNEDISNLTVKCPEEKCSFECRLIDMDEHLK-VCDNKTEKCPFFGYG 427 E + T+ +H+ L V CP + D+ HLK C+ CPF G Sbjct: 199 TKEFVFDTIQSHQYQ--CPRLPVACPNQCGVGTVAREDLPGHLKDSCNTALVLCPFKDSG 256 Query: 428 CTHKIKSKNMNSHMLREAQDHNNMLIDLLGNLKNEVIDLREK 553 C H+ M H+ + H M+ L+ + E+ +LR + Sbjct: 257 CKHRCPKLAMARHVEESVKPHLAMMCALVSRQRQELQELRRE 298
Score = 44.3 bits (103), Expect = 4e-04
Identities = 32/134 (23%), Positives = 58/134 (43%), Gaps = 3/134 (2%)
Frame = +2
Query: 77 DHKFSVIGEITNDMKCTLCGN-IAVNAQQALCGCNFCNKCITNFINNGGSACSGNTEECK 253
D+KF + + + C LCG + Q + CG FC+ C+ F++ G C E+
Sbjct: 5 DYKF--LEKPKRRLLCPLCGKPMREPVQVSTCGHRFCDTCLQEFLSEGVFKC---PEDQL 59
Query: 254 LENISQTVINHRLNEDISNLTVKC--PEEKCSFECRLIDMDEHLKVCDNKTEKCPFFGYG 427
+ ++ + L + L ++C EE C + L + HL C CP
Sbjct: 60 PLDYAKIYPDPELEVQVLGLPIRCIHSEEGCRWSGPLRHLQGHLNTCSFNVIPCP---NR 116
Query: 428 CTHKIKSKNMNSHM 469
C K+ +++ +H+
Sbjct: 117 CPMKLSRRDLPAHL 130
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 83,085,501
Number of Sequences: 369166
Number of extensions: 1582913
Number of successful extensions: 5746
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5287
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5702
length of database: 68,354,980
effective HSP length: 109
effective length of database: 48,218,865
effective search space used: 7763237265
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail