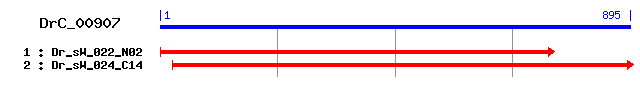
DrC_00907
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00907 (895 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P32908|SMC1_YEAST Structural maintenance of chromosome 1... 37 0.097 sp|Q00400|DTH1_DUGTI Homeobox protein DTH-1 36 0.17 sp|Q99N50|SYTL2_MOUSE Synaptotagmin-like protein 2 (Exophil... 34 0.48 sp|Q8DHW6|SYC_SYNEL Cysteinyl-tRNA synthetase (Cysteine--tR... 34 0.63 sp|Q03714|USA1_YEAST U1 SNP1-associating protein 1 33 0.82 sp|Q96YR5|RAD50_SULTO DNA double-strand break repair rad50 ... 33 0.82 sp|P51816|AFF2_HUMAN AF4/FMR2 family member 2 (Fragile X me... 33 1.1 sp|Q9HCH5|SYTL2_HUMAN Synaptotagmin-like protein 2 (Exophil... 33 1.1 sp|Q09958|EBP2_CAEEL Probable rRNA processing protein EBP2 ... 32 1.8 sp|P46954|SIP4_YEAST SIP4 protein 32 2.4
>sp|P32908|SMC1_YEAST Structural maintenance of chromosome 1 (DA-box protein SMC1) Length = 1225 Score = 36.6 bits (83), Expect = 0.097 Identities = 43/191 (22%), Positives = 73/191 (38%) Frame = +2 Query: 23 NYKLFNVCIKTLSNAKFLNTSVYIYTNRILYRSNILKGTDKNNSEDSNEIINLSNDSMSL 202 NY F + L L+ IL + DK ++ E N D Sbjct: 374 NYDKFKLNENDLKTYNCLHEKYLTEGGSILEEKIAVLNNDKREIQEELERFNKRADISKR 433 Query: 203 NIPKVNSVDLQKFDFENESFQVAVNERSTIDLSSSMHANLPKPPSEAVEINDRPYDSIVS 382 I + S+ +K D + +V++NE++ + + +H L K S+ N++ YD Sbjct: 434 RITEELSITGEKLDTQLNDLRVSLNEKNALH-TERLH-ELKKLQSDIESANNQEYDLNFK 491 Query: 383 LGEKYQKNDKFFSEELLDAVKKDNITHLQHLLSEERKLWLYFSNEKRFNIKISADAADLI 562 L E VK D+++ Q +ERKL + KRF + DL Sbjct: 492 LRETL--------------VKIDDLSANQRETMKERKLRENIAMLKRFFPGVKGLVHDLC 537 Query: 563 SQRRLNAGIAL 595 ++ G+A+ Sbjct: 538 HPKKEKYGLAV 548
>sp|Q00400|DTH1_DUGTI Homeobox protein DTH-1 Length = 533 Score = 35.8 bits (81), Expect = 0.17 Identities = 51/205 (24%), Positives = 82/205 (40%), Gaps = 19/205 (9%) Frame = +2 Query: 23 NYKLFNVCIKTLSNAKFLNTSVYIYTNRILYRSNILKGTDKNNSEDSNEIINLSNDSMSL 202 +YKL IKT+SN + + L RS N D I NL Sbjct: 264 SYKLSLPEIKTISNDWIKGSQNVNFNQNQLLRST--------NVSDYTLIKNLPQ----- 310 Query: 203 NIPKVNSVDLQKFDFENESFQVAVNERSTI---DLSSSMHANLPKPPSEAVEINDRPYDS 373 N+P N D NE+ Q N S + S +H N + +VE ND S Sbjct: 311 NLPNPNQTDSIYSSSINENNQPIRNYDSPNPDREDDSEIHENPNPHDTSSVENNDNENSS 370 Query: 374 IVSLGEKYQKNDKFFSEELLD------------AVKKDNITHLQHLLSEERKLWLYFSNE 517 +G+K ++ F +++L+ A +++++ +L L + K+W F N Sbjct: 371 SGDIGKKRKRRVLFSKKQILELERHFRQKKYLSAPEREHLANLIGLSPTQVKIW--FQNH 428 Query: 518 ----KRFNIKISADAADLISQRRLN 580 KR + + + + +L RRLN Sbjct: 429 RYKMKRAHHEKALEMGNLAVNRRLN 453
>sp|Q99N50|SYTL2_MOUSE Synaptotagmin-like protein 2 (Exophilin-4) Length = 950 Score = 34.3 bits (77), Expect = 0.48 Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 8/81 (9%) Frame = +2 Query: 431 LDAVKKDNITHLQHLLSEERKL------WLYFSNEKRFNIKISADAADLI--SQRRLNAG 586 L +++ + HL + ++++L W Y + KR KI AD+I S RR Sbjct: 25 LKRAEEERVRHLPEKIKDDQQLKNMSGQWFYEAKAKRHRDKIHG--ADIIRASMRRKKLP 82 Query: 587 IALEYQYDIMKQVTEYWLNNI 649 A E D + E W+NN+ Sbjct: 83 AAAEQNKDTAMRAKESWVNNV 103
>sp|Q8DHW6|SYC_SYNEL Cysteinyl-tRNA synthetase (Cysteine--tRNA ligase) (CysRS) Length = 486 Score = 33.9 bits (76), Expect = 0.63 Identities = 28/126 (22%), Positives = 51/126 (40%) Frame = +2 Query: 188 DSMSLNIPKVNSVDLQKFDFENESFQVAVNERSTIDLSSSMHANLPKPPSEAVEINDRPY 367 D M+L + + + + DF E+ A T+ + +H +P PP +A E+ P Sbjct: 294 DPMALRLLVLQAQYRKPLDFTPEALTAAAKGWQTLGEALHLHQQIPLPPIDAAEVRSHP- 352 Query: 368 DSIVSLGEKYQKNDKFFSEELLDAVKKDNITHLQHLLSEERKLWLYFSNEKRFNIKISAD 547 K + + E+L A I L L+ E+ +L+ R ++S D Sbjct: 353 --------KTEAFCQAMDEDLNTAAALAVIFELAKTLNREQHRYLHGGGWGRSPAEVSRD 404 Query: 548 AADLIS 565 L++ Sbjct: 405 WHTLVT 410
>sp|Q03714|USA1_YEAST U1 SNP1-associating protein 1 Length = 838 Score = 33.5 bits (75), Expect = 0.82 Identities = 26/88 (29%), Positives = 42/88 (47%), Gaps = 4/88 (4%) Frame = +2 Query: 74 LNTSVYIYTNRILYRSNILKGTDKNNSEDSNEIINLSNDSMSLNIPKVNSVDLQKFDFEN 253 +NT Y R L + +LK ++ DSN I L + + P ++ DL+ +D E Sbjct: 74 INTYFLSYEGRELSATCLLKDITSSSHPDSNHFIRLQLEKRT--SPSGSAFDLE-YDMEG 130 Query: 254 E----SFQVAVNERSTIDLSSSMHANLP 325 E + Q +N S+ + +SM NLP Sbjct: 131 EFNSMNIQFEINTLSSQRIFNSMEPNLP 158
>sp|Q96YR5|RAD50_SULTO DNA double-strand break repair rad50 ATPase Length = 879 Score = 33.5 bits (75), Expect = 0.82 Identities = 33/126 (26%), Positives = 59/126 (46%), Gaps = 2/126 (1%) Frame = +2 Query: 125 ILKGTDKNNSEDSNEIINLSNDSMSLNIPKVNSVDLQKFDFENESFQVAVNERSTIDLSS 304 ILK DSN I D ++ I ++ S++ K + EN+ Q E+ ++ Sbjct: 156 ILKIDKIEKLRDSNGPIKEVMDKINNKIIELQSLEKYKNESENQKIQ---KEKELENIKR 212 Query: 305 SMHANLPKPPSEAVEINDRPYDSIVSLGEKYQKNDKFFSE--ELLDAVKKDNITHLQHLL 478 + K E + Y+ IV L E+ +K +K + E LL+ + KD+I+ L+ + Sbjct: 213 ELEDLNIKEEKER-----KKYEDIVKLNEEEEKKEKRYVELISLLNKL-KDDISELREEV 266 Query: 479 SEERKL 496 +E +L Sbjct: 267 KDENRL 272
>sp|P51816|AFF2_HUMAN AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Ox19 protein) (Fragile X E mental retardation syndrome protein) Length = 1311 Score = 33.1 bits (74), Expect = 1.1 Identities = 26/87 (29%), Positives = 37/87 (42%) Frame = +2 Query: 95 YTNRILYRSNILKGTDKNNSEDSNEIINLSNDSMSLNIPKVNSVDLQKFDFENESFQVAV 274 YTN+ +N ++ T N E N + N SN + + IPK NSV + SF Sbjct: 60 YTNKGDALANRVQNTLGNYDEMKNLLTNHSNQNHLVGIPK-NSVPQNPNNKNEPSFFPEQ 118 Query: 275 NERSTIDLSSSMHANLPKPPSEAVEIN 355 R + H + P PP V +N Sbjct: 119 KNRIIPPHQDNTHPSAPMPPPSVVILN 145
>sp|Q9HCH5|SYTL2_HUMAN Synaptotagmin-like protein 2 (Exophilin-4) Length = 895 Score = 33.1 bits (74), Expect = 1.1 Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 8/81 (9%) Frame = +2 Query: 431 LDAVKKDNITHLQHLLSEERKL------WLYFSNEKRFNIKISADAADLI--SQRRLNAG 586 L +++ + HL + ++++L W Y + KR KI AD+I S R+ Sbjct: 10 LKRAEEERVRHLPEKIKDDQQLKNMSGQWFYEAKAKRHRDKIHG--ADIIRASMRKKRPQ 67 Query: 587 IALEYQYDIMKQVTEYWLNNI 649 IA E D E W+NN+ Sbjct: 68 IAAEQSKDRENGAKESWVNNV 88
>sp|Q09958|EBP2_CAEEL Probable rRNA processing protein EBP2 homolog Length = 340 Score = 32.3 bits (72), Expect = 1.8 Identities = 29/120 (24%), Positives = 53/120 (44%), Gaps = 9/120 (7%) Frame = +2 Query: 317 NLPKPPSEAVEINDRPYDSIVSLGEKYQKNDKFFSEELLDAVKKDNITHLQHL------- 475 N K +AV+I Y +++LG K + +++E K D TH+Q + Sbjct: 132 NFYKQAEKAVQI---AYPRLLNLGIKVLRPTDYYAE----MAKSD--THMQKVRKRLLGI 182 Query: 476 --LSEERKLWLYFSNEKRFNIKISADAADLISQRRLNAGIALEYQYDIMKQVTEYWLNNI 649 + E ++ + EK+F +K+ + + + N A++ MKQ E LNN+ Sbjct: 183 QEMKERQEAFRRIREEKKFAVKVQKEVLAAKNTEKKNLAEAVKKHKKGMKQQLEDMLNNV 242
>sp|P46954|SIP4_YEAST SIP4 protein Length = 829 Score = 32.0 bits (71), Expect = 2.4 Identities = 31/115 (26%), Positives = 60/115 (52%), Gaps = 5/115 (4%) Frame = +2 Query: 38 NVCIKTLSNAKFLNTSVYIYTNRILYRS--NILKGTDKNNSEDSNEIINLSNDSMSLNIP 211 N+ + SN+ FL+ S Y + N+I + N TD+ +ED +E + S++S+ L IP Sbjct: 685 NLLQHSSSNSNFLDASPYDF-NKIFMNNFENYDYETDEGYAEDDDEEDSDSDNSLPLEIP 743 Query: 212 KVNSVDLQKFDFENESFQVAVNER---STIDLSSSMHANLPKPPSEAVEINDRPY 367 S + K + S ++++ E +++D ++ + NL P S +V + + Y Sbjct: 744 FKKSKNKCKNRNKELSQRLSLFENRDSNSVDFNTDTNLNL-NPDSPSVTSSKKKY 797
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 78,309,689
Number of Sequences: 369166
Number of extensions: 1361013
Number of successful extensions: 4457
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4274
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4454
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 8982382310
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail