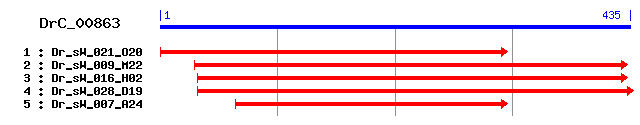
DrC_00863
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00863 (435 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q03344|ATIF1_RAT ATPase inhibitor, mitochondrial precursor 74 1e-13 sp|O35143|ATIF1_MOUSE ATPase inhibitor, mitochondrial precu... 74 1e-13 sp|O44441|ATIF2_CAEEL Probable ATPase inhibitor B0546.1, mi... 72 4e-13 sp|Q9UII2|ATIF1_HUMAN ATPase inhibitor, mitochondrial precu... 67 2e-11 sp|P01096|ATIF1_BOVIN ATPase inhibitor, mitochondrial precu... 67 2e-11 sp|Q29307|ATIF1_PIG ATPase inhibitor, mitochondrial precursor 65 8e-11 sp|P37209|ATIF1_CAEEL ATPase inhibitor, mitochondrial homolog 56 3e-08 sp|Q15643|TRIPB_HUMAN Thyroid receptor-interacting protein ... 35 0.090 sp|O61308|PUMA_PARUN 227 kDa spindle- and centromere-associ... 32 0.45 sp|P14105|MYH9_CHICK Myosin-9 (Myosin heavy chain, nonmuscl... 32 0.58
>sp|Q03344|ATIF1_RAT ATPase inhibitor, mitochondrial precursor Length = 107 Score = 74.3 bits (181), Expect = 1e-13 Identities = 33/66 (50%), Positives = 49/66 (74%) Frame = +2 Query: 149 TIRDAGGSFGKKEAYLEEEYFRKLNNRQIKELKDHLEDEIDTHTHEIEKHEEAVKRHKRK 328 +IR+AGG+FGK+E E+ YFR+ Q+ LK H EDEID H+ EIE+ ++ ++RHK+K Sbjct: 39 SIREAGGAFGKREKAEEDRYFREKTREQLAALKKHHEDEIDHHSKEIERLQKQIERHKKK 98 Query: 329 IEEIKN 346 I+ +KN Sbjct: 99 IKYLKN 104
>sp|O35143|ATIF1_MOUSE ATPase inhibitor, mitochondrial precursor Length = 106 Score = 74.3 bits (181), Expect = 1e-13 Identities = 32/66 (48%), Positives = 49/66 (74%) Frame = +2 Query: 149 TIRDAGGSFGKKEAYLEEEYFRKLNNRQIKELKDHLEDEIDTHTHEIEKHEEAVKRHKRK 328 +IR+AGG+FGK+E E+ YFR+ Q+ L+ H EDEID H+ EIE+ ++ + RHK+K Sbjct: 39 SIREAGGAFGKREKAEEDRYFREKTKEQLAALRKHHEDEIDHHSKEIERLQKQIDRHKKK 98 Query: 329 IEEIKN 346 I+++KN Sbjct: 99 IQQLKN 104
>sp|O44441|ATIF2_CAEEL Probable ATPase inhibitor B0546.1, mitochondrial precursor Length = 109 Score = 72.4 bits (176), Expect = 4e-13 Identities = 29/65 (44%), Positives = 50/65 (76%) Frame = +2 Query: 149 TIRDAGGSFGKKEAYLEEEYFRKLNNRQIKELKDHLEDEIDTHTHEIEKHEEAVKRHKRK 328 +IRDAGG+FGK EA E+EYF K Q++EL++H+++E+ H ++E H++ ++RH+++ Sbjct: 36 SIRDAGGAFGKMEAAREDEYFYKKQKAQLQELREHIQEEVKHHEGQLENHKKVLERHQQR 95 Query: 329 IEEIK 343 I EI+ Sbjct: 96 ISEIE 100
>sp|Q9UII2|ATIF1_HUMAN ATPase inhibitor, mitochondrial precursor sp|Q5RFJ9|ATIF1_PONPY ATPase inhibitor, mitochondrial precursor Length = 106 Score = 67.0 bits (162), Expect = 2e-11 Identities = 31/66 (46%), Positives = 47/66 (71%) Frame = +2 Query: 149 TIRDAGGSFGKKEAYLEEEYFRKLNNRQIKELKDHLEDEIDTHTHEIEKHEEAVKRHKRK 328 +IR+AGG+FGK+E EE YFR + Q+ LK H E+EI H EIE+ ++ ++RHK+K Sbjct: 39 SIREAGGAFGKREQAEEERYFRAQSREQLAALKKHHEEEIVHHKKEIERLQKEIERHKQK 98 Query: 329 IEEIKN 346 I+ +K+ Sbjct: 99 IKMLKH 104
>sp|P01096|ATIF1_BOVIN ATPase inhibitor, mitochondrial precursor Length = 109 Score = 67.0 bits (162), Expect = 2e-11 Identities = 31/69 (44%), Positives = 45/69 (65%) Frame = +2 Query: 152 IRDAGGSFGKKEAYLEEEYFRKLNNRQIKELKDHLEDEIDTHTHEIEKHEEAVKRHKRKI 331 +RDAGG+FGK+E EE YFR Q+ LK H E+EI H EIE+ ++ ++RHK+ I Sbjct: 40 VRDAGGAFGKREQAEEERYFRARAKEQLAALKKHHENEISHHAKEIERLQKEIERHKQSI 99 Query: 332 EEIKNLVKD 358 +++K D Sbjct: 100 KKLKQSEDD 108
>sp|Q29307|ATIF1_PIG ATPase inhibitor, mitochondrial precursor Length = 108 Score = 64.7 bits (156), Expect = 8e-11 Identities = 30/65 (46%), Positives = 43/65 (66%) Frame = +2 Query: 152 IRDAGGSFGKKEAYLEEEYFRKLNNRQIKELKDHLEDEIDTHTHEIEKHEEAVKRHKRKI 331 +R AGG+FGKKE EE YFR Q+ LK H E+ I H EIE+ ++ ++RHK+ I Sbjct: 40 VRXAGGAFGKKEQADEERYFRARAREQLAALKKHHENXISHHVKEIERLQKEIERHKQSI 99 Query: 332 EEIKN 346 +++KN Sbjct: 100 KKLKN 104
>sp|P37209|ATIF1_CAEEL ATPase inhibitor, mitochondrial homolog Length = 88 Score = 56.2 bits (134), Expect = 3e-08 Identities = 26/65 (40%), Positives = 42/65 (64%) Frame = +2 Query: 149 TIRDAGGSFGKKEAYLEEEYFRKLNNRQIKELKDHLEDEIDTHTHEIEKHEEAVKRHKRK 328 +IR+AGGS G A EEEYFR+ Q+ LK LE ++ EI HE+ +++H+++ Sbjct: 19 SIREAGGSLGMMGATREEEYFRRQQKDQLDNLKKKLEADMTQSQQEIRDHEKVLEQHQQR 78 Query: 329 IEEIK 343 ++EI+ Sbjct: 79 LKEIE 83
>sp|Q15643|TRIPB_HUMAN Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution related gene on chromosome 14) Length = 1979 Score = 34.7 bits (78), Expect = 0.090 Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 15/56 (26%) Frame = +2 Query: 233 IKELKDHLEDEIDTHTHEI---------------EKHEEAVKRHKRKIEEIKNLVK 355 IKELK + EID H HE+ +H E + ++ +IEE++NL++ Sbjct: 218 IKELKQNRSQEIDDHQHEMSVLQNAHQQKLTEISRRHREELSDYEERIEELENLLQ 273
>sp|O61308|PUMA_PARUN 227 kDa spindle- and centromere-associated protein (PUMA1) Length = 1955 Score = 32.3 bits (72), Expect = 0.45 Identities = 19/70 (27%), Positives = 35/70 (50%) Frame = +2 Query: 158 DAGGSFGKKEAYLEEEYFRKLNNRQIKELKDHLEDEIDTHTHEIEKHEEAVKRHKRKIEE 337 D G + G+ + E + NR+ K + L+ +IDT + E E V+R K+KI + Sbjct: 1070 DLGNATGRLRSSEEHNAILQSENRKSKTEIEALKHQIDTIMNTKESCESEVERLKKKIVQ 1129 Query: 338 IKNLVKDHDK 367 + K+ ++ Sbjct: 1130 TTTITKEQNE 1139
>sp|P14105|MYH9_CHICK Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A) Length = 1959 Score = 32.0 bits (71), Expect = 0.58 Identities = 16/60 (26%), Positives = 32/60 (53%), Gaps = 4/60 (6%) Frame = +2 Query: 200 EEYFRKLNNRQIKELKDHLEDEIDTHTHEI----EKHEEAVKRHKRKIEEIKNLVKDHDK 367 ++ R +++ LK LEDE TH +I +KH +A++ ++E+ K + + +K Sbjct: 1158 QQELRSKREQEVTVLKKTLEDEAKTHEAQIQEMRQKHSQAIEELAEQLEQTKRVKANLEK 1217
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,018,525
Number of Sequences: 369166
Number of extensions: 449514
Number of successful extensions: 2796
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 2530
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2771
length of database: 68,354,980
effective HSP length: 100
effective length of database: 49,881,480
effective search space used: 2194785120
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail