DrC_00754
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00754 (1491 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O19049|HNRPK_RABIT Heterogeneous nuclear ribonucleoprote... 112 4e-24 sp|P61980|HNRPK_RAT Heterogeneous nuclear ribonucleoprotein... 110 8e-24 sp|P60335|PCBP1_MOUSE Poly(rC)-binding protein 1 (Alpha-CP1... 79 4e-14 sp|Q15366|PCBP2_HUMAN Poly(rC)-binding protein 2 (Alpha-CP2... 77 2e-13 sp|Q61990|PCBP2_MOUSE Poly(rC)-binding protein 2 (Alpha-CP2... 74 8e-13 sp|P57722|PCBP3_MOUSE Poly(rC)-binding protein 3 (Alpha-CP3) 71 7e-12 sp|P57721|PCBP3_HUMAN Poly(rC)-binding protein 3 (Alpha-CP3) 71 7e-12 sp|P57724|PCBP4_MOUSE Poly(rC)-binding protein 4 (Alpha-CP4) 61 1e-08 sp|P57723|PCBP4_HUMAN Poly(rC)-binding protein 4 (Alpha-CP4) 60 1e-08 sp|Q96I24|FUBP3_HUMAN Far upstream element-binding protein ... 60 2e-08
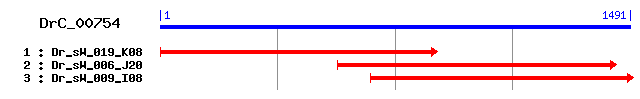
>sp|O19049|HNRPK_RABIT Heterogeneous nuclear ribonucleoprotein K (hnRNP K) Length = 463 Score = 112 bits (279), Expect = 4e-24 Identities = 63/192 (32%), Positives = 105/192 (54%), Gaps = 13/192 (6%) Frame = +1 Query: 82 IEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGPERVFILESTMDNCIKVWH 261 +E R L+ SK A +IGKGG+ I+ LR + NA +S+PD+ GPER+ + + ++ ++ Sbjct: 43 VELRILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSGPERILSISADIETIGEILK 102 Query: 262 DCLPDLKDCM---FPNVDQRKSSGS----------FRDDNPDMDLRFLVHDSQVGAIIGK 402 +P L++ + P + S F+ + D +LR L+H S G IIG Sbjct: 103 KIIPTLEEGLQLPSPTATSQLPLESDAVECLNYQHFKGSDFDCELRLLIHQSLAGGIIGV 162 Query: 403 GGVNVKNLRMQYQLNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFRVIYETLESQSIKGK 582 G +K LR Q IK+F P+S+DR+V + GK ++V EC ++I + + IKG+ Sbjct: 163 KGAKIKELRENTQ-TTIKLFQECCPHSTDRVVLIGGKPDRVVECIKIILDLISESPIKGR 221 Query: 583 IENYNSENFDES 618 + Y+ +DE+ Sbjct: 222 AQPYDPNFYDET 233
Score = 59.7 bits (143), Expect = 2e-08
Identities = 31/67 (46%), Positives = 44/67 (65%)
Frame = +1
Query: 1219 QVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETILSLTGTKDQIDGAQYL 1398
QV+IP L S+ GK G+ + I +SGA I ID E + + I+++TGT+DQI AQYL
Sbjct: 391 QVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQDQIQNAQYL 450
Query: 1399 LQICIKK 1419
LQ +K+
Sbjct: 451 LQNSVKQ 457
Score = 47.4 bits (111), Expect = 1e-04
Identities = 31/93 (33%), Positives = 46/93 (49%), Gaps = 4/93 (4%)
Frame = +1
Query: 295 PNVDQRKSSGSFRDDNPD--MDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQLNVIKVFPV 468
P D + R N D ++LR L+ GA+IGKGG N+K LR Y + V
Sbjct: 23 PAEDMEEEQAFKRSRNTDEMVELRILLQSKNAGAVIGKGGKNIKALRTDYNAS------V 76
Query: 469 TAPNSS--DRLVQMVGKFEKVFECFRVIYETLE 561
+ P+SS +R++ + E + E + I TLE
Sbjct: 77 SVPDSSGPERILSISADIETIGEILKKIIPTLE 109
Score = 40.0 bits (92), Expect = 0.018
Identities = 22/58 (37%), Positives = 31/58 (53%), Gaps = 2/58 (3%)
Frame = +1
Query: 73 GNRIEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSI--PDNHGPERVFILESTMD 240
G I + IP A IIGKGG+RI+++RHE A + I P +R+ + T D
Sbjct: 385 GPIITTQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQD 442
>sp|P61980|HNRPK_RAT Heterogeneous nuclear ribonucleoprotein K (dC stretch-binding protein) (CSBP) sp|P61979|HNRPK_MOUSE Heterogeneous nuclear ribonucleoprotein K sp|P61978|HNRPK_HUMAN Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP) Length = 463 Score = 110 bits (276), Expect = 8e-24 Identities = 62/192 (32%), Positives = 105/192 (54%), Gaps = 13/192 (6%) Frame = +1 Query: 82 IEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGPERVFILESTMDNCIKVWH 261 +E R L+ SK A +IGKGG+ I+ LR + NA +S+PD+ GPER+ + + ++ ++ Sbjct: 43 VELRILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSGPERILSISADIETIGEILK 102 Query: 262 DCLPDLKDCM---FPNVDQRKSSGS----------FRDDNPDMDLRFLVHDSQVGAIIGK 402 +P L++ + P + S ++ + D +LR L+H S G IIG Sbjct: 103 KIIPTLEEGLQLPSPTATSQLPLESDAVECLNYQHYKGSDFDCELRLLIHQSLAGGIIGV 162 Query: 403 GGVNVKNLRMQYQLNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFRVIYETLESQSIKGK 582 G +K LR Q IK+F P+S+DR+V + GK ++V EC ++I + + IKG+ Sbjct: 163 KGAKIKELRENTQ-TTIKLFQECCPHSTDRVVLIGGKPDRVVECIKIILDLISESPIKGR 221 Query: 583 IENYNSENFDES 618 + Y+ +DE+ Sbjct: 222 AQPYDPNFYDET 233
Score = 59.7 bits (143), Expect = 2e-08
Identities = 31/67 (46%), Positives = 44/67 (65%)
Frame = +1
Query: 1219 QVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETILSLTGTKDQIDGAQYL 1398
QV+IP L S+ GK G+ + I +SGA I ID E + + I+++TGT+DQI AQYL
Sbjct: 391 QVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQDQIQNAQYL 450
Query: 1399 LQICIKK 1419
LQ +K+
Sbjct: 451 LQNSVKQ 457
Score = 47.4 bits (111), Expect = 1e-04
Identities = 31/93 (33%), Positives = 46/93 (49%), Gaps = 4/93 (4%)
Frame = +1
Query: 295 PNVDQRKSSGSFRDDNPD--MDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQLNVIKVFPV 468
P D + R N D ++LR L+ GA+IGKGG N+K LR Y + V
Sbjct: 23 PAEDMEEEQAFKRSRNTDEMVELRILLQSKNAGAVIGKGGKNIKALRTDYNAS------V 76
Query: 469 TAPNSS--DRLVQMVGKFEKVFECFRVIYETLE 561
+ P+SS +R++ + E + E + I TLE
Sbjct: 77 SVPDSSGPERILSISADIETIGEILKKIIPTLE 109
Score = 40.0 bits (92), Expect = 0.018
Identities = 22/58 (37%), Positives = 31/58 (53%), Gaps = 2/58 (3%)
Frame = +1
Query: 73 GNRIEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSI--PDNHGPERVFILESTMD 240
G I + IP A IIGKGG+RI+++RHE A + I P +R+ + T D
Sbjct: 385 GPIITTQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQD 442
>sp|P60335|PCBP1_MOUSE Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) sp|Q15365|PCBP1_HUMAN Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid-binding protein SUB2.3) sp|O19048|PCBP1_RABIT Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) Length = 356 Score = 78.6 bits (192), Expect = 4e-14 Identities = 55/172 (31%), Positives = 91/172 (52%), Gaps = 4/172 (2%) Frame = +1 Query: 82 IEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGPERVFILESTMDNCIKVWH 261 + R L+ K IIGK GE ++R+R E AR++I + + PER+ L + K + Sbjct: 14 LTIRLLMHGKEVGSIIGKKGESVKRIREESGARINISEGNCPERIITLTGPTNAIFKAFA 73 Query: 262 DCLPDLKDCMFPNVDQRKSSGSFRDDNPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQ 441 + L++ + N S+ + R P + LR +V +Q G++IGKGG +K +R Sbjct: 74 MIIDKLEEDI--NSSMTNSTAASR---PPVTLRLVVPATQCGSLIGKGGCKIKEIRESTG 128 Query: 442 LNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFR----VIYETLESQSIKGKI 585 V +V PNS++R + + G + V EC + V+ ETL SQS +G++ Sbjct: 129 AQV-QVAGDMLPNSTERAITIAGVPQSVTECVKQICLVMLETL-SQSPQGRV 178
Score = 44.7 bits (104), Expect = 7e-04
Identities = 22/74 (29%), Positives = 41/74 (55%)
Frame = +1
Query: 340 NPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQLNVIKVFPVTAPNSSDRLVQMVGKFE 519
N + +R L+H +VG+IIGK G +VK +R + + ++ N +R++ + G
Sbjct: 11 NVTLTIRLLMHGKEVGSIIGKKGESVKRIREESGARI----NISEGNCPERIITLTGPTN 66
Query: 520 KVFECFRVIYETLE 561
+F+ F +I + LE
Sbjct: 67 AIFKAFAMIIDKLE 80
Score = 36.6 bits (83), Expect = 0.19
Identities = 24/72 (33%), Positives = 41/72 (56%), Gaps = 1/72 (1%)
Frame = +1
Query: 1189 ASSNASIEKL-QVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETILSLTG 1365
AS +AS + +++IP +LI + G+ G ++ I SGA+I I E + +++TG
Sbjct: 272 ASLDASTQTTHELTIPNNLIGCIIGRQGANINEIRQMSGAQIKIANPVEGSSGRQVTITG 331
Query: 1366 TKDQIDGAQYLL 1401
+ I AQYL+
Sbjct: 332 SAASISLAQYLI 343
>sp|Q15366|PCBP2_HUMAN Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2) Length = 365 Score = 76.6 bits (187), Expect = 2e-13 Identities = 50/170 (29%), Positives = 85/170 (50%), Gaps = 4/170 (2%) Frame = +1 Query: 82 IEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGPERVFILESTMDNCIKVWH 261 + R L+ K IIGK GE ++++R E AR++I + + PER+ L + K + Sbjct: 14 LTIRLLMHGKEVGSIIGKKGESVKKMREESGARINISEGNCPERIITLAGPTNAIFKAFA 73 Query: 262 DCLPDLKDCMFPNVDQRKSSGSFRDDNPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQ 441 + L++ + ++ + S P + LR +V SQ G++IGKGG +K +R Sbjct: 74 MIIDKLEEDISSSM-----TNSTAASRPPVTLRLVVPASQCGSLIGKGGCKIKEIRESTG 128 Query: 442 LNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFR----VIYETLESQSIKG 579 V +V PNS++R + + G + + EC + V+ ETL KG Sbjct: 129 AQV-QVAGDMLPNSTERAITIAGIPQSIIECVKQICVVMLETLSQSPPKG 177
Score = 44.7 bits (104), Expect = 7e-04
Identities = 22/74 (29%), Positives = 41/74 (55%)
Frame = +1
Query: 340 NPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQLNVIKVFPVTAPNSSDRLVQMVGKFE 519
N + +R L+H +VG+IIGK G +VK +R + + ++ N +R++ + G
Sbjct: 11 NVTLTIRLLMHGKEVGSIIGKKGESVKKMREESGARI----NISEGNCPERIITLAGPTN 66
Query: 520 KVFECFRVIYETLE 561
+F+ F +I + LE
Sbjct: 67 AIFKAFAMIIDKLE 80
Score = 40.8 bits (94), Expect = 0.010
Identities = 22/74 (29%), Positives = 40/74 (54%)
Frame = +1
Query: 1186 GASSNASIEKLQVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETILSLTG 1365
G ++A +++IP LI + G+ G ++ I SGA+I I E + + +++TG
Sbjct: 280 GLDASAQTTSHELTIPNDLIGCIIGRQGAKINEIRQMSGAQIKIANPVEGSTDRQVTITG 339
Query: 1366 TKDQIDGAQYLLQI 1407
+ I AQYL+ +
Sbjct: 340 SAASISLAQYLINV 353
>sp|Q61990|PCBP2_MOUSE Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP) Length = 362 Score = 74.3 bits (181), Expect = 8e-13 Identities = 47/166 (28%), Positives = 82/166 (49%) Frame = +1 Query: 82 IEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGPERVFILESTMDNCIKVWH 261 + R L+ K IIGK GE ++++R E AR++I + + PER+ L + K + Sbjct: 14 LTIRLLMHGKEVGSIIGKKGESVKKMREESGARINISEGNCPERIITLAGPTNAIFKAFA 73 Query: 262 DCLPDLKDCMFPNVDQRKSSGSFRDDNPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQ 441 + L++ + ++ + S P + LR +V SQ G++IGKGG +K +R Sbjct: 74 MIIDKLEEDISSSM-----TNSTAASRPPVTLRLVVPASQCGSLIGKGGCKIKEIRESTG 128 Query: 442 LNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFRVIYETLESQSIKG 579 V +V PNS++R + + G + + EC + I + KG Sbjct: 129 AQV-QVAGDMLPNSTERAITIAGIPQSIIECVKQICVVMLESPPKG 173
Score = 44.7 bits (104), Expect = 7e-04
Identities = 22/74 (29%), Positives = 41/74 (55%)
Frame = +1
Query: 340 NPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQLNVIKVFPVTAPNSSDRLVQMVGKFE 519
N + +R L+H +VG+IIGK G +VK +R + + ++ N +R++ + G
Sbjct: 11 NVTLTIRLLMHGKEVGSIIGKKGESVKKMREESGARI----NISEGNCPERIITLAGPTN 66
Query: 520 KVFECFRVIYETLE 561
+F+ F +I + LE
Sbjct: 67 AIFKAFAMIIDKLE 80
Score = 40.8 bits (94), Expect = 0.010
Identities = 22/74 (29%), Positives = 40/74 (54%)
Frame = +1
Query: 1186 GASSNASIEKLQVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETILSLTG 1365
G ++A +++IP LI + G+ G ++ I SGA+I I E + + +++TG
Sbjct: 277 GLDASAQTTSHELTIPNDLIGCIIGRQGAKINEIRQMSGAQIKIANPVEGSTDRQVTITG 336
Query: 1366 TKDQIDGAQYLLQI 1407
+ I AQYL+ +
Sbjct: 337 SAASISLAQYLINV 350
>sp|P57722|PCBP3_MOUSE Poly(rC)-binding protein 3 (Alpha-CP3) Length = 339 Score = 71.2 bits (173), Expect = 7e-12 Identities = 47/166 (28%), Positives = 80/166 (48%) Frame = +1 Query: 82 IEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGPERVFILESTMDNCIKVWH 261 + R L+ K IIGK GE ++++R E AR++I + + PER+ + D K + Sbjct: 14 LTIRLLMHGKEVGSIIGKKGETVKKMREESGARINISEGNCPERIVTITGPTDAIFKAFA 73 Query: 262 DCLPDLKDCMFPNVDQRKSSGSFRDDNPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQ 441 ++ + ++ S S P + LR +V SQ G++IGKGG +K +R Sbjct: 74 MIAYKFEEDIINSM-----SNSPATSKPPVTLRLVVPASQCGSLIGKGGSKIKEIRESTG 128 Query: 442 LNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFRVIYETLESQSIKG 579 V +V PNS++R V + G + + +C + I + KG Sbjct: 129 AQV-QVAGDMLPNSTERAVTISGTPDAIIQCVKQICVVMLESPPKG 173
Score = 42.4 bits (98), Expect = 0.004
Identities = 23/78 (29%), Positives = 40/78 (51%)
Frame = +1
Query: 340 NPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQLNVIKVFPVTAPNSSDRLVQMVGKFE 519
N + +R L+H +VG+IIGK G VK +R + + ++ N +R+V + G +
Sbjct: 11 NVTLTIRLLMHGKEVGSIIGKKGETVKKMREESGARI----NISEGNCPERIVTITGPTD 66
Query: 520 KVFECFRVIYETLESQSI 573
+F+ F +I E I
Sbjct: 67 AIFKAFAMIAYKFEEDII 84
Score = 42.4 bits (98), Expect = 0.004
Identities = 28/82 (34%), Positives = 42/82 (51%)
Frame = +1
Query: 1189 ASSNASIEKLQVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETILSLTGT 1368
AS AS +L +IP LI + G+ G ++ I SGA+I I E + E +++TGT
Sbjct: 257 ASPPASTHEL--TIPNDLIGCIIGRQGTKINEIRQMSGAQIKIANATEGSSERQITITGT 314
Query: 1369 KDQIDGAQYLLQICIKKSVLNL 1434
I AQYL+ + V +
Sbjct: 315 PANISLAQYLINARLTSEVTGM 336
>sp|P57721|PCBP3_HUMAN Poly(rC)-binding protein 3 (Alpha-CP3) Length = 339 Score = 71.2 bits (173), Expect = 7e-12 Identities = 47/166 (28%), Positives = 80/166 (48%) Frame = +1 Query: 82 IEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGPERVFILESTMDNCIKVWH 261 + R L+ K IIGK GE ++++R E AR++I + + PER+ + D K + Sbjct: 14 LTIRLLMHGKEVGSIIGKKGETVKKMREESGARINISEGNCPERIVTITGPTDAIFKAFA 73 Query: 262 DCLPDLKDCMFPNVDQRKSSGSFRDDNPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQ 441 ++ + ++ S S P + LR +V SQ G++IGKGG +K +R Sbjct: 74 MIAYKFEEDIINSM-----SNSPATSKPPVTLRLVVPASQCGSLIGKGGSKIKEIRESTG 128 Query: 442 LNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFRVIYETLESQSIKG 579 V +V PNS++R V + G + + +C + I + KG Sbjct: 129 AQV-QVAGDMLPNSTERAVTISGTPDAIIQCVKQICVVMLESPPKG 173
Score = 42.4 bits (98), Expect = 0.004
Identities = 23/78 (29%), Positives = 40/78 (51%)
Frame = +1
Query: 340 NPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQLNVIKVFPVTAPNSSDRLVQMVGKFE 519
N + +R L+H +VG+IIGK G VK +R + + ++ N +R+V + G +
Sbjct: 11 NVTLTIRLLMHGKEVGSIIGKKGETVKKMREESGARI----NISEGNCPERIVTITGPTD 66
Query: 520 KVFECFRVIYETLESQSI 573
+F+ F +I E I
Sbjct: 67 AIFKAFAMIAYKFEEDII 84
Score = 42.4 bits (98), Expect = 0.004
Identities = 28/82 (34%), Positives = 42/82 (51%)
Frame = +1
Query: 1189 ASSNASIEKLQVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETILSLTGT 1368
AS AS +L +IP LI + G+ G ++ I SGA+I I E + E +++TGT
Sbjct: 257 ASPPASTHEL--TIPNDLIGCIIGRQGTKINEIRQMSGAQIKIANATEGSSERQITITGT 314
Query: 1369 KDQIDGAQYLLQICIKKSVLNL 1434
I AQYL+ + V +
Sbjct: 315 PANISLAQYLINARLTSEVTGM 336
>sp|P57724|PCBP4_MOUSE Poly(rC)-binding protein 4 (Alpha-CP4) Length = 403 Score = 60.8 bits (146), Expect = 1e-08 Identities = 50/177 (28%), Positives = 80/177 (45%), Gaps = 5/177 (2%) Frame = +1 Query: 82 IEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGPERVFILESTMDNCIKVWH 261 + R L+ K IIGK GE ++R+R + +AR++I + PER+ + + V+H Sbjct: 18 LTLRMLMHGKEVGSIIGKKGETVKRIREQSSARITISEGSCPERITTITG---STAAVFH 74 Query: 262 DCLP-----DLKDCMFPNVDQRKSSGSFRDDNPDMDLRFLVHDSQVGAIIGKGGVNVKNL 426 D C P + GS P + LR ++ SQ G++IGK G +K + Sbjct: 75 AVSMIAFKLDEDLCAAP-----ANGGSV--SRPPVTLRLVIPASQCGSLIGKAGTKIKEI 127 Query: 427 RMQYQLNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFRVIYETLESQSIKGKIENYN 597 R V +V PNS++R V + G + + C R I + KG Y+ Sbjct: 128 RETTGAQV-QVAGDLLPNSTERAVTVSGVPDAIILCVRQICAVILESPPKGATIPYH 183
Score = 42.4 bits (98), Expect = 0.004
Identities = 25/82 (30%), Positives = 38/82 (46%)
Frame = +1
Query: 1171 PDNIVGASSNASIEKLQVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETI 1350
P + G + + +P LI V G+ G +S I SGA I I E E
Sbjct: 229 PSVVPGMDPSTQTSSQEFLVPNDLIGCVIGRQGSKISEIRQMSGAHIKIGNQAEGAGERH 288
Query: 1351 LSLTGTKDQIDGAQYLLQICIK 1416
+++TG+ I AQYL+ C++
Sbjct: 289 VTITGSPVSIALAQYLITACLE 310
>sp|P57723|PCBP4_HUMAN Poly(rC)-binding protein 4 (Alpha-CP4) Length = 403 Score = 60.5 bits (145), Expect = 1e-08 Identities = 45/172 (26%), Positives = 80/172 (46%) Frame = +1 Query: 82 IEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGPERVFILESTMDNCIKVWH 261 + R L+ K IIGK GE ++R+R + +AR++I + PER+ +T+ Sbjct: 18 LTLRMLMHGKEVGSIIGKKGETVKRIREQSSARITISEGSCPERI----TTITGSTAAVF 73 Query: 262 DCLPDLKDCMFPNVDQRKSSGSFRDDNPDMDLRFLVHDSQVGAIIGKGGVNVKNLRMQYQ 441 + + + ++ ++G P + LR ++ SQ G++IGK G +K +R Sbjct: 74 HAVSMIAFKLDEDLCAAPANGG-NVSRPPVTLRLVIPASQCGSLIGKAGTKIKEIRETTG 132 Query: 442 LNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFRVIYETLESQSIKGKIENYN 597 V +V PNS++R V + G + + C R I + KG Y+ Sbjct: 133 AQV-QVAGDLLPNSTERAVTVSGVPDAIILCVRQICAVILESPPKGATIPYH 183
Score = 42.0 bits (97), Expect = 0.005
Identities = 25/82 (30%), Positives = 37/82 (45%)
Frame = +1
Query: 1171 PDNIVGASSNASIEKLQVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETI 1350
P + G + +P LI V G+ G +S I SGA I I E E
Sbjct: 229 PSVVPGLDPGTQTSSQEFLVPNDLIGCVIGRQGSKISEIRQMSGAHIKIGNQAEGAGERH 288
Query: 1351 LSLTGTKDQIDGAQYLLQICIK 1416
+++TG+ I AQYL+ C++
Sbjct: 289 VTITGSPVSIALAQYLITACLE 310
>sp|Q96I24|FUBP3_HUMAN Far upstream element-binding protein 3 (FUSE-binding protein 3) Length = 572 Score = 60.1 bits (144), Expect = 2e-08 Identities = 42/176 (23%), Positives = 82/176 (46%), Gaps = 7/176 (3%) Frame = +1 Query: 94 FLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHG--PERVFILESTMDNCIKVWHDC 267 F +P K IIG+GGE+I R++ E ++ I PER +L T ++ Sbjct: 82 FKVPDKMVGFIIGRGGEQISRIQAESGCKIQIASESSGIPERPCVLTGTPES-------- 133 Query: 268 LPDLKDCMFPNVDQRKSSGSFRDDNPDMDL--RFLVHDSQVGAIIGKGGVNVKNLRMQYQ 441 + K + VD+ ++ F +D L+ S+VG +IG+GG +K L+ + Sbjct: 134 IEQAKRLLGQIVDRCRNGPGFHNDIDSNSTIQEILIPASKVGLVIGRGGETIKQLQERTG 193 Query: 442 LNVIKVFPVTAPNSSDRLVQMVGKFEKVFECFRVIYETL---ESQSIKGKIENYNS 600 + ++ + P +D+ +++ G KV + ++ E + + +G ++NS Sbjct: 194 VKMVMIQDGPLPTGADKPLRITGDAFKVQQAREMVLEIIREKDQADFRGVRGDFNS 249
Score = 42.7 bits (99), Expect = 0.003
Identities = 30/124 (24%), Positives = 53/124 (42%), Gaps = 12/124 (9%)
Frame = +1
Query: 100 IPSKAASIIIGKGGERIQRLRHECNARLSIPDNHG--PERVFILESTMDNCIKVWHDCLP 273
+P A I+IG+ GE I++++++ R+ + G PER + D C H
Sbjct: 260 VPRFAVGIVIGRNGEMIKKIQNDAGVRIQFKPDDGISPERAAQVMGPPDRCQHAAHIISE 319
Query: 274 DLKDCM----FPNVDQRKSSGSFRDD------NPDMDLRFLVHDSQVGAIIGKGGVNVKN 423
+ F + + G R D ++ + V + G +IGKGG N+K+
Sbjct: 320 LILTAQERDGFGGLAAARGRGRGRGDWSVGAPGGVQEITYTVPADKCGLVIGKGGENIKS 379
Query: 424 LRMQ 435
+ Q
Sbjct: 380 INQQ 383
Score = 36.6 bits (83), Expect = 0.19
Identities = 15/47 (31%), Positives = 25/47 (53%)
Frame = +1
Query: 67 APGNRIEFRFLIPSKAASIIIGKGGERIQRLRHECNARLSIPDNHGP 207
APG E + +P+ ++IGKGGE I+ + + A + + N P
Sbjct: 350 APGGVQEITYTVPADKCGLVIGKGGENIKSINQQSGAHVELQRNPPP 396
Score = 36.2 bits (82), Expect = 0.25
Identities = 19/61 (31%), Positives = 32/61 (52%)
Frame = +1
Query: 1219 QVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETILSLTGTKDQIDGAQYL 1398
+ +P ++ + G+ GE +S I ++SG +I I E LTGT + I+ A+ L
Sbjct: 81 EFKVPDKMVGFIIGRGGEQISRIQAESGCKIQIASESSGIPERPCVLTGTPESIEQAKRL 140
Query: 1399 L 1401
L
Sbjct: 141 L 141
Score = 36.2 bits (82), Expect = 0.25
Identities = 17/62 (27%), Positives = 33/62 (53%)
Frame = +1
Query: 1216 LQVSIPTSLIHSVKGKNGEVLSNIISDSGAEISIDGNDEQNMETILSLTGTKDQIDGAQY 1395
++VS+P + V G+NGE++ I +D+G I +D + E + G D+ A +
Sbjct: 256 IEVSVPRFAVGIVIGRNGEMIKKIQNDAGVRIQFKPDDGISPERAAQVMGPPDRCQHAAH 315
Query: 1396 LL 1401
++
Sbjct: 316 II 317
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 158,673,213
Number of Sequences: 369166
Number of extensions: 3105234
Number of successful extensions: 9658
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7983
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9086
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 17949083355
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail