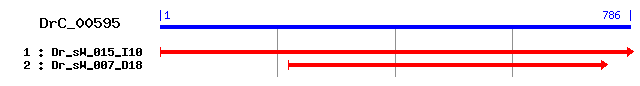
DrC_00595
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00595 (786 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P21195|PDIA1_RABIT Protein disulfide-isomerase precursor... 81 4e-15 sp|P07237|PDIA1_HUMAN Protein disulfide-isomerase precursor... 78 2e-14 sp|Q5R5B6|PDIA1_PONPY Protein disulfide-isomerase precursor... 78 2e-14 sp|Q9XI01|PDI1_ARATH Probable protein disulfide-isomerase 1... 77 4e-14 sp|P54399|PDI_DROME Protein disulfide-isomerase precursor (... 77 7e-14 sp|P80284|PDI_HORVU Protein disulfide-isomerase precursor (... 77 7e-14 sp|P04785|PDIA1_RAT Protein disulfide-isomerase precursor (... 76 9e-14 sp|Q5I0H9|PDIA5_RAT Protein disulfide-isomerase A5 precursor 76 9e-14 sp|Q8R4U2|PDIA1_CRIGR Protein disulfide-isomerase precursor... 76 9e-14 sp|P13667|PDIA4_HUMAN Protein disulfide-isomerase A4 precur... 76 1e-13
>sp|P21195|PDIA1_RABIT Protein disulfide-isomerase precursor (PDI) (Prolyl 4-hydroxylase beta subunit) (Cellular thyroid hormone binding protein) (p55) Length = 509 Score = 80.9 bits (198), Expect = 4e-15 Identities = 45/109 (41%), Positives = 64/109 (58%), Gaps = 3/109 (2%) Frame = +1 Query: 175 VVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDA 354 V++L++SNF + + + K V FY+PWCGHC A+ P AA KL +A VDA Sbjct: 27 VLVLKSSNFAEELAAHKH-LLVEFYAPWCGHCKALAPEYAKAAGKLKAEGSDIRLAKVDA 85 Query: 355 TSETNLAAQYKVTGFPTIIYFSPSGDWV---KYVGERTEDAIVQFMMKK 492 T E++LA QY V G+PTI +F +GD +Y R D IV ++ K+ Sbjct: 86 TEESDLAQQYGVRGYPTIKFFK-NGDTASPKEYTAGREADDIVNWLKKR 133
Score = 60.8 bits (146), Expect = 4e-09
Identities = 42/138 (30%), Positives = 64/138 (46%), Gaps = 5/138 (3%)
Frame = +1
Query: 103 FMTNPPKPKLDARR---HWRRIPGHEAVVILETSNFVQNIVSQKSGSFVMFYSPWCGHCV 273
F+ KP L ++ W R P V +L NF + +K FV FY+PWCGHC
Sbjct: 347 FLEGKIKPHLMSQELPEDWDRQP----VKVLVGKNFEEVAFDEKKNVFVEFYAPWCGHCK 402
Query: 274 AMKPALVNAANKLHNLLPSCLVAAVDATSETNLAAQYKVTGFPTIIYF--SPSGDWVKYV 447
+ P + + + + ++A +D+T+ N KV FPT+ +F P + Y
Sbjct: 403 QLAP-IWDKLGETYKEHQDIVIAKMDSTA--NEVEAVKVHSFPTLKFFPAGPGRTVIDYN 459
Query: 448 GERTEDAIVQFMMKKDSD 501
GERT D +F+ D
Sbjct: 460 GERTLDGFKKFLESGGQD 477
>sp|P07237|PDIA1_HUMAN Protein disulfide-isomerase precursor (PDI) (Prolyl 4-hydroxylase beta subunit) (Cellular thyroid hormone binding protein) (p55) Length = 508 Score = 78.2 bits (191), Expect = 2e-14 Identities = 45/109 (41%), Positives = 62/109 (56%), Gaps = 3/109 (2%) Frame = +1 Query: 175 VVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDA 354 V++L SNF + + + K V FY+PWCGHC A+ P AA KL +A VDA Sbjct: 26 VLVLRKSNFAEALAAHKY-LLVEFYAPWCGHCKALAPEYAKAAGKLKAEGSEIRLAKVDA 84 Query: 355 TSETNLAAQYKVTGFPTIIYFSPSGDWV---KYVGERTEDAIVQFMMKK 492 T E++LA QY V G+PTI +F +GD +Y R D IV ++ K+ Sbjct: 85 TEESDLAQQYGVRGYPTIKFFR-NGDTASPKEYTAGREADDIVNWLKKR 132
Score = 61.6 bits (148), Expect = 2e-09
Identities = 45/151 (29%), Positives = 70/151 (46%), Gaps = 5/151 (3%)
Frame = +1
Query: 64 EYSLEREEDDFIAFMTNPPKPKLDARR---HWRRIPGHEAVVILETSNFVQNIVSQKSGS 234
E + ER + F+ KP L ++ W + P V +L NF +K
Sbjct: 333 ELTAERITEFCHRFLEGKIKPHLMSQELPEDWDKQP----VKVLVGKNFEDVAFDEKKNV 388
Query: 235 FVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDATSETNLAAQYKVTGFPTIIY 414
FV FY+PWCGHC + P + + + + + ++A +D+T+ N KV FPT+ +
Sbjct: 389 FVEFYAPWCGHCKQLAP-IWDKLGETYKDHENIVIAKMDSTA--NEVEAVKVHSFPTLKF 445
Query: 415 FSPSGD--WVKYVGERTEDAIVQFMMKKDSD 501
F S D + Y GERT D +F+ D
Sbjct: 446 FPASADRTVIDYNGERTLDGFKKFLESGGQD 476
>sp|Q5R5B6|PDIA1_PONPY Protein disulfide-isomerase precursor (PDI) (Prolyl 4-hydroxylase beta subunit) Length = 508 Score = 78.2 bits (191), Expect = 2e-14 Identities = 45/109 (41%), Positives = 62/109 (56%), Gaps = 3/109 (2%) Frame = +1 Query: 175 VVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDA 354 V++L SNF + + + K V FY+PWCGHC A+ P AA KL +A VDA Sbjct: 26 VLVLRKSNFAEALAAHKY-LLVEFYAPWCGHCKALAPEYAKAAGKLKAEGSEIRLAKVDA 84 Query: 355 TSETNLAAQYKVTGFPTIIYFSPSGDWV---KYVGERTEDAIVQFMMKK 492 T E++LA QY V G+PTI +F +GD +Y R D IV ++ K+ Sbjct: 85 TEESDLAQQYGVRGYPTIKFFR-NGDTASPKEYTAGREADDIVNWLKKR 132
Score = 61.6 bits (148), Expect = 2e-09
Identities = 45/151 (29%), Positives = 70/151 (46%), Gaps = 5/151 (3%)
Frame = +1
Query: 64 EYSLEREEDDFIAFMTNPPKPKLDARR---HWRRIPGHEAVVILETSNFVQNIVSQKSGS 234
E + ER + F+ KP L ++ W + P V +L NF +K
Sbjct: 333 ELTAERITEFCHRFLEGKIKPHLMSQELPDDWDKQP----VKVLVGKNFEDVAFDEKKNV 388
Query: 235 FVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDATSETNLAAQYKVTGFPTIIY 414
FV FY+PWCGHC + P + + + + + ++A +D+T+ N KV FPT+ +
Sbjct: 389 FVEFYAPWCGHCKQLAP-IWDKLGETYKDHENIVIAKMDSTA--NEVEAVKVHSFPTLKF 445
Query: 415 FSPSGD--WVKYVGERTEDAIVQFMMKKDSD 501
F S D + Y GERT D +F+ D
Sbjct: 446 FPASADRTVIDYNGERTLDGFKKFLESGGQD 476
>sp|Q9XI01|PDI1_ARATH Probable protein disulfide-isomerase 1 precursor (PDI 1) Length = 501 Score = 77.4 bits (189), Expect = 4e-14 Identities = 45/118 (38%), Positives = 65/118 (55%), Gaps = 3/118 (2%) Frame = +1 Query: 169 EAVVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAV 348 E V+ L+ +NF I ++ V FY+PWCGHC + P AA+ L + +P ++A + Sbjct: 30 EFVLTLDHTNFTDTI-NKHDFIVVEFYAPWCGHCKQLAPEYEKAASALSSNVPPVVLAKI 88 Query: 349 DATSETN--LAAQYKVTGFPTIIYFSPSGDWV-KYVGERTEDAIVQFMMKKDSDLKYE 513 DA+ ETN A QY+V GFPTI F G V +Y G R + IV ++ K+ E Sbjct: 89 DASEETNREFATQYEVQGFPTIKIFRNGGKAVQEYNGpreAEGIVTYLKKQSGPASAE 146
Score = 65.1 bits (157), Expect = 2e-10
Identities = 38/122 (31%), Positives = 63/122 (51%), Gaps = 4/122 (3%)
Frame = +1
Query: 166 HEAVVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAA 345
+E V ++ + + +++ + FY+PWCGHC + P L A + S ++A
Sbjct: 373 NEPVKVVVSDSLDDIVLNSGKNVLLEFYAPWCGHCQKLAPILDEVAVSYQSD-SSVVIAK 431
Query: 346 VDATSETNLAAQYKVTGFPTIIYFSPSGDWVKYVGERTEDAIVQFMMKKDSDL----KYE 513
+DAT+ + V GFPTI + S SG+ V Y G+RT++ + F+ K + K E
Sbjct: 432 LDATANDFPKDTFDVKGFPTIYFKSASGNVVVYEGDRTKEDFISFVDKNKDTVGEPKKEE 491
Query: 514 ET 519
ET
Sbjct: 492 ET 493
>sp|P54399|PDI_DROME Protein disulfide-isomerase precursor (PDI) Length = 496 Score = 76.6 bits (187), Expect = 7e-14 Identities = 43/108 (39%), Positives = 59/108 (54%) Frame = +1 Query: 169 EAVVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAV 348 E V++ NF Q +++ V FY+PWCGHC A+ P AA +L +A V Sbjct: 27 EGVLVATVDNFKQ-LIADNEFVLVEFYAPWCGHCKALAPEYAKAAQQLAEKESPIKLAKV 85 Query: 349 DATSETNLAAQYKVTGFPTIIYFSPSGDWVKYVGERTEDAIVQFMMKK 492 DAT E LA QY V G+PT+ +F SG V+Y G R I+ ++ KK Sbjct: 86 DATVEGELAEQYAVRGYPTLKFFR-SGSPVEYSGGRQAADIIAWVTKK 132
Score = 55.1 bits (131), Expect = 2e-07
Identities = 34/104 (32%), Positives = 53/104 (50%), Gaps = 1/104 (0%)
Frame = +1
Query: 175 VVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDA 354
V +L +SNF + + V FY+PWCGHC + P A K + ++A +D+
Sbjct: 369 VKVLVSSNFESVALDKSKSVLVEFYAPWCGHCKQLAPIYDQLAEKYKD-NEDIVIAKMDS 427
Query: 355 TSETNLAAQYKVTGFPTIIYFSPSGDWV-KYVGERTEDAIVQFM 483
T+ N K++ FPTI YF + V + +RT D V+F+
Sbjct: 428 TA--NELESIKISSFPTIKYFRKEDNKVIDFNLDRTLDDFVKFL 469
>sp|P80284|PDI_HORVU Protein disulfide-isomerase precursor (PDI) (Endosperm protein E-1) Length = 513 Score = 76.6 bits (187), Expect = 7e-14 Identities = 45/135 (33%), Positives = 69/135 (51%), Gaps = 3/135 (2%) Frame = +1 Query: 97 IAFMTNPPKPKLDARRHWRRIPGHEAVVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVA 276 +A + + P + + EAV+ L NF + + Q V FY+PWCGHC + Sbjct: 14 LAVVLSAPAARAEEAAAAEEAAAPEAVLTLHADNF-DDAIGQHPFILVEFYAPWCGHCKS 72 Query: 277 MKPALVNAANKLHNLLPSCLVAAVDATSETN--LAAQYKVTGFPTIIYFSPSGDWV-KYV 447 + P AA L P+ ++A VDA E N LA +Y+V GFPT+ F G + +Y Sbjct: 73 LAPEYEKAAQLLSKHDPAIVLAKVDANDEKNKPLAGKYEVQGFPTLKIFRNGGKSIQEYK 132 Query: 448 GERTEDAIVQFMMKK 492 G R + IV+++ K+ Sbjct: 133 GpreAEGIVEYLKKQ 147
Score = 73.2 bits (178), Expect = 8e-13
Identities = 44/138 (31%), Positives = 69/138 (50%), Gaps = 2/138 (1%)
Frame = +1
Query: 82 EEDDFIAFMTNPPKPKLDARRHWRRIP--GHEAVVILETSNFVQNIVSQKSGSFVMFYSP 255
E +A++ + KL R IP +E V ++ N + + FY+P
Sbjct: 350 EAGQIVAWLKDYFDGKLTPFRKSEPIPEANNEPVKVVVADNVHDVVFKSGKNVLIEFYAP 409
Query: 256 WCGHCVAMKPALVNAANKLHNLLPSCLVAAVDATSETNLAAQYKVTGFPTIIYFSPSGDW 435
WCGHC + P L AA L + ++A +DAT E ++ ++ V G+PT+ + +PSG
Sbjct: 410 WCGHCKKLAPILDEAAATLQSE-EDVVIAKMDAT-ENDVPGEFDVQGYPTLYFVTPSGKK 467
Query: 436 VKYVGERTEDAIVQFMMK 489
V Y G RT D IV ++ K
Sbjct: 468 VSYEGGRTADEIVDYIRK 485
>sp|P04785|PDIA1_RAT Protein disulfide-isomerase precursor (PDI) (Prolyl 4-hydroxylase beta subunit) (Cellular thyroid hormone binding protein) Length = 509 Score = 76.3 bits (186), Expect = 9e-14 Identities = 44/109 (40%), Positives = 62/109 (56%), Gaps = 3/109 (2%) Frame = +1 Query: 175 VVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDA 354 V++L+ SNF + + + V FY+PWCGHC A+ P AA KL +A VDA Sbjct: 28 VLVLKKSNFAEALAAHNY-LLVEFYAPWCGHCKALAPEYAKAAAKLKAEGSEIRLAKVDA 86 Query: 355 TSETNLAAQYKVTGFPTIIYFSPSGDWV---KYVGERTEDAIVQFMMKK 492 T E++LA QY V G+PTI +F +GD +Y R D IV ++ K+ Sbjct: 87 TEESDLAQQYGVRGYPTIKFFK-NGDTASPKEYTAGREADDIVNWLKKR 134
Score = 61.2 bits (147), Expect = 3e-09
Identities = 42/138 (30%), Positives = 66/138 (47%), Gaps = 5/138 (3%)
Frame = +1
Query: 103 FMTNPPKPKLDARR---HWRRIPGHEAVVILETSNFVQNIVSQKSGSFVMFYSPWCGHCV 273
F+ KP L ++ W + P V +L NF + +K FV FY+PWCGHC
Sbjct: 348 FLEGKIKPHLMSQELPEDWDKQP----VKVLVGKNFEEVAFDEKKNVFVEFYAPWCGHCK 403
Query: 274 AMKPALVNAANKLHNLLPSCLVAAVDATSETNLAAQYKVTGFPTIIYFSPSGD--WVKYV 447
+ P + + + + + ++A +D+T+ N KV FPT+ +F S D + Y
Sbjct: 404 QLAP-IWDKLGETYKDHENIVIAKMDSTA--NEVEAVKVHSFPTLKFFPASADRTVIDYN 460
Query: 448 GERTEDAIVQFMMKKDSD 501
GERT D +F+ D
Sbjct: 461 GERTLDGFKKFLESGGQD 478
>sp|Q5I0H9|PDIA5_RAT Protein disulfide-isomerase A5 precursor Length = 517 Score = 76.3 bits (186), Expect = 9e-14 Identities = 53/168 (31%), Positives = 82/168 (48%), Gaps = 2/168 (1%) Frame = +1 Query: 22 IFRYFNYGKNTSYSEYSLEREEDDFIAFMTNPPKPKLDA-RRHWRRIPGHEAVVILETSN 198 +F+Y NYG +D + ++ NP P+ W G +V L + Sbjct: 237 LFQYENYGSTA-----------EDIVEWLKNPQPPQPQVPETPWADEGG--SVYHLTDED 283 Query: 199 FVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLH-NLLPSCLVAAVDATSETNLA 375 F Q V + S VMF++PWCGHC MKP +AA LH + S ++AAVDAT LA Sbjct: 284 FDQ-FVKEHSSVLVMFHAPWCGHCKKMKPEFESAAEVLHGDAESSGVLAAVDATINEALA 342 Query: 376 AQYKVTGFPTIIYFSPSGDWVKYVGERTEDAIVQFMMKKDSDLKYEET 519 ++ ++ FPT+ YF +G+ RT+ +++M ++ E T Sbjct: 343 ERFHISAFPTLKYFK-NGEQQAVPALRTKKKFIEWMQNPEAPPPPEPT 389
Score = 59.3 bits (142), Expect = 1e-08
Identities = 50/172 (29%), Positives = 73/172 (42%), Gaps = 3/172 (1%)
Frame = +1
Query: 19 PIFRYFNYGKNTSYSEYSLEREEDDFIAFMTNPPKPKLDARRHWRRIPGHEAVVILETSN 198
P +YF G+ + R + FI +M NP P W +V+ L N
Sbjct: 351 PTLKYFKNGEQQAVPAL---RTKKKFIEWMQNPEAPP-PPEPTWEE--QQTSVLHLVGDN 404
Query: 199 FVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDATSETN--L 372
F + + +K + VMFY+PWC HC + P A+ + AAVD + N L
Sbjct: 405 FRETL-KKKKHTLVMFYAPWCPHCKKVIPHFTATADAFKD-DRKIACAAVDCVKDKNQDL 462
Query: 373 AAQYKVTGFPTIIYFSPSGDWVKYVGERTEDAIVQFMMK-KDSDLKYEETQR 525
Q V +PT Y+ KY +RTE F+ ++ DLK E +R
Sbjct: 463 CQQESVKAYPTFHYYHYGKLVEKYESDRTELGFTSFIRTLREGDLKRLEKRR 514
Score = 58.9 bits (141), Expect = 2e-08
Identities = 41/158 (25%), Positives = 69/158 (43%), Gaps = 5/158 (3%)
Frame = +1
Query: 13 EDPIFRYFNYGKNTSYSEYSLEREEDDFIAFMTNPPKPKLDARRHWRRIPGHEAVVILET 192
+D F+Y + +Y +AF+ +P P L W PG + VV +++
Sbjct: 102 KDKKIELFHYQDGAFHMQYDRAVTLKSIVAFLKDPKGPPL-----WEEDPGAKDVVHIDS 156
Query: 193 S-NFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKL--HNLLPSCLVAAVDATSE 363
+F + + ++ +MFY+PWC C + P AA ++ H +L V +
Sbjct: 157 EKDFRRLLKKEEKPLLMMFYAPWCSMCKRIMPHFQKAATQVRGHTVLAGMNVYPPEF--- 213
Query: 364 TNLAAQYKVTGFPTIIYFSPSGDWVKY--VGERTEDAI 471
N+ +Y V G+PTI YF +Y G ED +
Sbjct: 214 ENIKEEYNVRGYPTICYFEKGRFLFQYENYGSTAEDIV 251
>sp|Q8R4U2|PDIA1_CRIGR Protein disulfide-isomerase precursor (PDI) (Prolyl 4-hydroxylase beta subunit) (p58) Length = 509 Score = 76.3 bits (186), Expect = 9e-14 Identities = 44/109 (40%), Positives = 62/109 (56%), Gaps = 3/109 (2%) Frame = +1 Query: 175 VVILETSNFVQNIVSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDA 354 V++L+ SNF + + + V FY+PWCGHC A+ P AA KL +A VDA Sbjct: 28 VLVLKKSNFAEALAAHNY-LLVEFYAPWCGHCKALAPEYAKAAAKLKAEGSEIRLAKVDA 86 Query: 355 TSETNLAAQYKVTGFPTIIYFSPSGDWV---KYVGERTEDAIVQFMMKK 492 T E++LA QY V G+PTI +F +GD +Y R D IV ++ K+ Sbjct: 87 TEESDLAQQYGVRGYPTIKFFK-NGDTASPKEYTAGREADDIVNWLKKR 134
Score = 60.5 bits (145), Expect = 5e-09
Identities = 41/138 (29%), Positives = 66/138 (47%), Gaps = 5/138 (3%)
Frame = +1
Query: 103 FMTNPPKPKLDARR---HWRRIPGHEAVVILETSNFVQNIVSQKSGSFVMFYSPWCGHCV 273
F+ KP L ++ W + P V +L NF + +K FV FY+PWCGHC
Sbjct: 348 FLEGKIKPHLMSQELPEDWDKQP----VKVLVGKNFEEVAFDEKKNVFVEFYAPWCGHCK 403
Query: 274 AMKPALVNAANKLHNLLPSCLVAAVDATSETNLAAQYKVTGFPTIIYFSPSGD--WVKYV 447
+ P + + + + + ++A +D+T+ N KV FPT+ +F + D + Y
Sbjct: 404 QLAP-IWDKLGETYKDHENIIIAKMDSTA--NEVEAVKVHSFPTLKFFPATADRTVIDYN 460
Query: 448 GERTEDAIVQFMMKKDSD 501
GERT D +F+ D
Sbjct: 461 GERTLDGFKKFLESGGQD 478
>sp|P13667|PDIA4_HUMAN Protein disulfide-isomerase A4 precursor (Protein ERp-72) (ERp72) Length = 645 Score = 75.9 bits (185), Expect = 1e-13 Identities = 46/143 (32%), Positives = 73/143 (51%) Frame = +1 Query: 64 EYSLEREEDDFIAFMTNPPKPKLDARRHWRRIPGHEAVVILETSNFVQNIVSQKSGSFVM 243 +Y R +++ +A + +P W P E ++L NF +V+ V Sbjct: 150 DYEGSRTQEEIVAKVREVSQPD------WT--PPPEVTLVLTKENF-DEVVNDADIILVE 200 Query: 244 FYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDATSETNLAAQYKVTGFPTIIYFSP 423 FY+PWCGHC + P AA +L P +A VDAT+ET+LA ++ V+G+PT+ F Sbjct: 201 FYAPWCGHCKKLAPEYEKAAKELSKRSPPIPLAKVDATAETDLAKRFDVSGYPTLKIFR- 259 Query: 424 SGDWVKYVGERTEDAIVQFMMKK 492 G Y G R + IV +M+++ Sbjct: 260 KGRPYDYNGpreKYGIVDYMIEQ 282
Score = 75.1 bits (183), Expect = 2e-13
Identities = 47/133 (35%), Positives = 64/133 (48%)
Frame = +1
Query: 76 EREEDDFIAFMTNPPKPKLDARRHWRRIPGHEAVVILETSNFVQNIVSQKSGSFVMFYSP 255
E EEDD D + V++L +NF N V+ K + FY+P
Sbjct: 44 EEEEDD-------------DEEEDDLEVKEENGVLVLNDANF-DNFVADKDTVLLEFYAP 89
Query: 256 WCGHCVAMKPALVNAANKLHNLLPSCLVAAVDATSETNLAAQYKVTGFPTIIYFSPSGDW 435
WCGHC P AN L + P VA +DATS + LA+++ V+G+PTI G
Sbjct: 90 WCGHCKQFAPEYEKIANILKDKDPPIPVAKIDATSASVLASRFDVSGYPTIKILK-KGQA 148
Query: 436 VKYVGERTEDAIV 474
V Y G RT++ IV
Sbjct: 149 VDYEGSRTQEEIV 161
Score = 67.0 bits (162), Expect = 6e-11
Identities = 43/133 (32%), Positives = 71/133 (53%), Gaps = 7/133 (5%)
Frame = +1
Query: 55 SYSEYSLEREEDD-------FIAFMTNPPKPKLDARRHWRRIPGHEAVVILETSNFVQNI 213
S ++++E EE D AF KP + ++ + G VV+ +T + + +
Sbjct: 482 SGKKFAMEPEEFDSDTLREFVTAFKKGKLKPVIKSQPVPKNNKGPVKVVVGKTFDSI--V 539
Query: 214 VSQKSGSFVMFYSPWCGHCVAMKPALVNAANKLHNLLPSCLVAAVDATSETNLAAQYKVT 393
+ K + FY+PWCGHC ++P + N+ K + ++A +DAT+ + +YKV
Sbjct: 540 MDPKKDVLIEFYAPWCGHCKQLEP-VYNSLAKKYKGQKGLVIAKMDATANDVPSDRYKVE 598
Query: 394 GFPTIIYFSPSGD 432
GFPT IYF+PSGD
Sbjct: 599 GFPT-IYFAPSGD 610
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,754,594
Number of Sequences: 369166
Number of extensions: 1372370
Number of successful extensions: 3628
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3365
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3522
length of database: 68,354,980
effective HSP length: 108
effective length of database: 48,403,600
effective search space used: 7405750800
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail