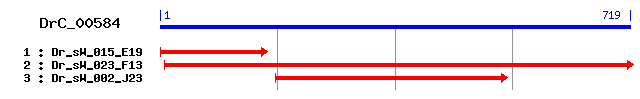
DrC_00584
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00584 (719 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q8MHZ9|CL46_BOVIN Collectin-46 precursor (CL-46) (46 kDa... 78 3e-14 sp|Q28008|CLC3A_BOVIN C-type lectin domain family 3 member ... 72 1e-12 sp|P05452|TETN_HUMAN Tetranectin precursor (TN) (Plasminoge... 71 3e-12 sp|P43025|TETN_MOUSE TETRANECTIN preCURSOR (TN) (PLASMINOGE... 71 3e-12 sp|P26258|TETN_CARSP Tetranectin-like protein 70 4e-12 sp|P42916|CL43_BOVIN Collectin-43 precursor (CL-43) (43 kDa... 70 6e-12 sp|P23805|CONG_BOVIN Conglutinin precursor 69 2e-11 sp|O75596|CLC3A_HUMAN C-type lectin domain family 3 member ... 69 2e-11 sp|P81282|CSPG2_BOVIN Versican core protein precursor (Larg... 68 2e-11 sp|O02659|MBL2_BOVIN Mannose-binding protein C precursor (M... 68 2e-11
>sp|Q8MHZ9|CL46_BOVIN Collectin-46 precursor (CL-46) (46 kDa collectin) Length = 371 Score = 77.8 bits (190), Expect = 3e-14 Identities = 43/108 (39%), Positives = 58/108 (53%), Gaps = 3/108 (2%) Frame = +1 Query: 292 TYNEALQFCRTRGTQLARVNSASDNQAVASFAKRVVADYYWIDANDYTEEGKWVDSNGKE 471 +Y++A Q CR QLA SA++N+AVA + D + + ND + EGK+ G+ Sbjct: 267 SYSDAQQLCREAKGQLASPRSAAENEAVAQLVRAKNNDAF-LSMNDISTEGKFTYPTGES 325 Query: 472 KPYKNWAAHEPNGNRA---ENCVFSFYLEEGTWRDISCYNKYLAICTF 606 Y NWA+ EPN N A ENCV + EG W D+ C L IC F Sbjct: 326 LVYSNWASGEPNNNNAGQPENCVQIY--REGKWNDVPCSEPLLVICEF 371
Score = 43.9 bits (102), Expect = 4e-04
Identities = 25/67 (37%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Frame = +1
Query: 19 WLDGNDAKTEGRWVDSNDNDLKHKGFHPGEP---NGGVNENCLNGYTYIDGSWNDFPCTY 189
+L ND TEG++ L + + GEP N G ENC+ Y +G WND PC+
Sbjct: 306 FLSMNDISTEGKFTYPTGESLVYSNWASGEPNNNNAGQPENCVQ--IYREGKWNDVPCS- 362
Query: 190 KNLYAIC 210
+ L IC
Sbjct: 363 EPLLVIC 369
>sp|Q28008|CLC3A_BOVIN C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin) Length = 197 Score = 72.0 bits (175), Expect = 1e-12 Identities = 35/107 (32%), Positives = 56/107 (52%), Gaps = 2/107 (1%) Frame = +1 Query: 295 YNEALQFCRTRGTQLARVNSASDNQAVASFAKRVV--ADYYWIDANDYTEEGKWVDSNGK 468 ++EA + C ++G L SA + A+ + KR + + +W+ ND EGK+VD NG Sbjct: 88 FHEANEDCISKGGTLVVPRSADEINALRDYGKRSLPGVNDFWLGINDMVAEGKFVDINGL 147 Query: 469 EKPYKNWAAHEPNGNRAENCVFSFYLEEGTWRDISCYNKYLAICTFS 609 + NW +PNG + ENC +G W D +C++ IC F+ Sbjct: 148 AISFLNWDQAQPNGGKRENCALFSQSAQGKWSDEACHSSKRYICEFT 194
Score = 45.1 bits (105), Expect = 2e-04
Identities = 23/65 (35%), Positives = 30/65 (46%)
Frame = +1
Query: 16 YWLDGNDAKTEGRWVDSNDNDLKHKGFHPGEPNGGVNENCLNGYTYIDGSWNDFPCTYKN 195
+WL ND EG++VD N + + +PNGG ENC G W+D C
Sbjct: 128 FWLGINDMVAEGKFVDINGLAISFLNWDQAQPNGGKRENCALFSQSAQGKWSDEACHSSK 187
Query: 196 LYAIC 210
Y IC
Sbjct: 188 RY-IC 191
>sp|P05452|TETN_HUMAN Tetranectin precursor (TN) (Plasminogen-kringle 4 binding protein) Length = 202 Score = 71.2 bits (173), Expect = 3e-12 Identities = 38/124 (30%), Positives = 59/124 (47%), Gaps = 5/124 (4%) Frame = +1 Query: 250 EVDSEKMIVSTDRYTYNEALQFCRTRGTQLARVNSASDNQAVASFAKRVVADY--YWIDA 423 +V + + T T++EA + C +RG L+ + S+N A+ + ++ V + W+ Sbjct: 76 KVHMKCFLAFTQTKTFHEASEDCISRGGTLSTPQTGSENDALYEYLRQSVGNEAEIWLGL 135 Query: 424 NDYTEEGKWVDSNGKEKPYKNWAAH---EPNGNRAENCVFSFYLEEGTWRDISCYNKYLA 594 ND EG WVD G YKNW +P+G + ENC G W D C ++ Sbjct: 136 NDMAAEGTWVDMTGARIAYKNWETEITAQPDGGKTENCAVLSGAANGKWFDKRCRDQLPY 195 Query: 595 ICTF 606 IC F Sbjct: 196 ICQF 199
Score = 38.9 bits (89), Expect = 0.014
Identities = 20/58 (34%), Positives = 26/58 (44%), Gaps = 3/58 (5%)
Frame = +1
Query: 19 WLDGNDAKTEGRWVDSNDNDLKHKGFH---PGEPNGGVNENCLNGYTYIDGSWNDFPC 183
WL ND EG WVD + +K + +P+GG ENC +G W D C
Sbjct: 132 WLGLNDMAAEGTWVDMTGARIAYKNWETEITAQPDGGKTENCAVLSGAANGKWFDKRC 189
>sp|P43025|TETN_MOUSE TETRANECTIN preCURSOR (TN) (PLASMINOGEN-KRINGLE 4 BINDING PROTEIN) Length = 202 Score = 71.2 bits (173), Expect = 3e-12 Identities = 39/125 (31%), Positives = 62/125 (49%), Gaps = 5/125 (4%) Frame = +1 Query: 250 EVDSEKMIVSTDRYTYNEALQFCRTRGTQLARVNSASDNQAVASFAKRVVAD--YYWIDA 423 +V+ + ++ T T++EA + C ++G L S +N+A+ +A+ V + W+ Sbjct: 76 KVNLKCLLAFTQPKTFHEASEDCISQGGTLGTPQSELENEALFEYARHSVGNDANIWLGL 135 Query: 424 NDYTEEGKWVDSNGKEKPYKNWAAH---EPNGNRAENCVFSFYLEEGTWRDISCYNKYLA 594 ND EG WVD G YKNW +P+G +AENC G W D C ++ Sbjct: 136 NDMAAEGAWVDMTGGLLAYKNWETEITTQPDGGKAENCAALSGAANGKWFDKRCRDQLPY 195 Query: 595 ICTFS 609 IC F+ Sbjct: 196 ICQFA 200
Score = 38.5 bits (88), Expect = 0.018
Identities = 21/58 (36%), Positives = 26/58 (44%), Gaps = 3/58 (5%)
Frame = +1
Query: 19 WLDGNDAKTEGRWVDSNDNDLKHKGFH---PGEPNGGVNENCLNGYTYIDGSWNDFPC 183
WL ND EG WVD L +K + +P+GG ENC +G W D C
Sbjct: 132 WLGLNDMAAEGAWVDMTGGLLAYKNWETEITTQPDGGKAENCAALSGAANGKWFDKRC 189
>sp|P26258|TETN_CARSP Tetranectin-like protein Length = 166 Score = 70.5 bits (171), Expect = 4e-12 Identities = 39/114 (34%), Positives = 59/114 (51%), Gaps = 2/114 (1%) Frame = +1 Query: 271 IVSTDRYTYNEALQFCRTRGTQLARVNSASDNQAVASFAKR--VVADYYWIDANDYTEEG 444 + S +Y+ A + C +G L+ S+ + ++ S+AK+ V A +WI ND T EG Sbjct: 49 LASRGSKSYHAANEDCIAQGGTLSIPRSSDEGNSLRSYAKKSLVGARDFWIGVNDMTTEG 108 Query: 445 KWVDSNGKEKPYKNWAAHEPNGNRAENCVFSFYLEEGTWRDISCYNKYLAICTF 606 K+VD NG Y NW +P G ENCV + +G W D C ++ IC + Sbjct: 109 KFVDVNGLPITYFNWDRSKPVGGTRENCVAASTSGQGKWSDDVCRSEKRYICEY 162
Score = 48.1 bits (113), Expect = 2e-05
Identities = 23/65 (35%), Positives = 34/65 (52%)
Frame = +1
Query: 16 YWLDGNDAKTEGRWVDSNDNDLKHKGFHPGEPNGGVNENCLNGYTYIDGSWNDFPCTYKN 195
+W+ ND TEG++VD N + + + +P GG ENC+ T G W+D C +
Sbjct: 97 FWIGVNDMTTEGKFVDVNGLPITYFNWDRSKPVGGTRENCVAASTSGQGKWSDDVCRSEK 156
Query: 196 LYAIC 210
Y IC
Sbjct: 157 RY-IC 160
>sp|P42916|CL43_BOVIN Collectin-43 precursor (CL-43) (43 kDa collectin) Length = 321 Score = 70.1 bits (170), Expect = 6e-12 Identities = 41/110 (37%), Positives = 58/110 (52%), Gaps = 5/110 (4%) Frame = +1 Query: 292 TYNEALQFCRTRGTQLARVNSASDNQAVASFAKRVVADYYWIDANDYTEEGKWVDSNGKE 471 +Y++A Q CR QLA S+++N+AV R + ++ ND ++EGK+ G Sbjct: 216 SYSDAEQLCREAKGQLASPRSSAENEAVTQLV-RAKNKHAYLSMNDISKEGKFTYPTGGS 274 Query: 472 KPYKNWAAHEPNGNRA-----ENCVFSFYLEEGTWRDISCYNKYLAICTF 606 Y NWA EPN NRA ENC+ + +G W DI C + L IC F Sbjct: 275 LDYSNWAPGEPN-NRAKDEGPENCLEIY--SDGNWNDIECREERLVICEF 321
Score = 43.9 bits (102), Expect = 4e-04
Identities = 25/68 (36%), Positives = 32/68 (47%), Gaps = 4/68 (5%)
Frame = +1
Query: 19 WLDGNDAKTEGRWVDSNDNDLKHKGFHPGEPNGGVN----ENCLNGYTYIDGSWNDFPCT 186
+L ND EG++ L + + PGEPN ENCL Y DG+WND C
Sbjct: 255 YLSMNDISKEGKFTYPTGGSLDYSNWAPGEPNNRAKDEGPENCLE--IYSDGNWNDIECR 312
Query: 187 YKNLYAIC 210
+ L IC
Sbjct: 313 EERL-VIC 319
>sp|P23805|CONG_BOVIN Conglutinin precursor Length = 371 Score = 68.6 bits (166), Expect = 2e-11 Identities = 38/108 (35%), Positives = 57/108 (52%), Gaps = 3/108 (2%) Frame = +1 Query: 292 TYNEALQFCRTRGTQLARVNSASDNQAVASFAKRVVADYYWIDANDYTEEGKWVDSNGKE 471 +Y++A Q CR QLA S+++N+AV + + Y + ND + EG++ G+ Sbjct: 267 SYSDAEQLCREAKGQLASPRSSAENEAVTQMVRAQEKNAY-LSMNDISTEGRFTYPTGEI 325 Query: 472 KPYKNWAAHEPNGN---RAENCVFSFYLEEGTWRDISCYNKYLAICTF 606 Y NWA EPN + + ENCV F +G W D+ C + L IC F Sbjct: 326 LVYSNWADGEPNNSDEGQPENCVEIF--PDGKWNDVPCSKQLLVICEF 371
Score = 46.6 bits (109), Expect = 7e-05
Identities = 27/67 (40%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Frame = +1
Query: 19 WLDGNDAKTEGRWVDSNDNDLKHKGFHPGEPNG---GVNENCLNGYTYIDGSWNDFPCTY 189
+L ND TEGR+ L + + GEPN G ENC+ + DG WND PC+
Sbjct: 306 YLSMNDISTEGRFTYPTGEILVYSNWADGEPNNSDEGQPENCVE--IFPDGKWNDVPCS- 362
Query: 190 KNLYAIC 210
K L IC
Sbjct: 363 KQLLVIC 369
>sp|O75596|CLC3A_HUMAN C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin) Length = 197 Score = 68.6 bits (166), Expect = 2e-11 Identities = 34/107 (31%), Positives = 56/107 (52%), Gaps = 2/107 (1%) Frame = +1 Query: 295 YNEALQFCRTRGTQLARVNSASDNQAVASFAKRVV--ADYYWIDANDYTEEGKWVDSNGK 468 ++EA + C ++G L ++ + A+ + KR + + +W+ ND EGK+VD NG Sbjct: 88 FHEANEDCISKGGILVIPRNSDEINALQDYGKRSLPGVNDFWLGINDMVTEGKFVDVNGI 147 Query: 469 EKPYKNWAAHEPNGNRAENCVFSFYLEEGTWRDISCYNKYLAICTFS 609 + NW +PNG + ENCV +G W D +C + IC F+ Sbjct: 148 AISFLNWDRAQPNGGKRENCVLFSQSAQGKWSDEACRSSKRYICEFT 194
Score = 47.8 bits (112), Expect = 3e-05
Identities = 24/65 (36%), Positives = 32/65 (49%)
Frame = +1
Query: 16 YWLDGNDAKTEGRWVDSNDNDLKHKGFHPGEPNGGVNENCLNGYTYIDGSWNDFPCTYKN 195
+WL ND TEG++VD N + + +PNGG ENC+ G W+D C
Sbjct: 128 FWLGINDMVTEGKFVDVNGIAISFLNWDRAQPNGGKRENCVLFSQSAQGKWSDEACRSSK 187
Query: 196 LYAIC 210
Y IC
Sbjct: 188 RY-IC 191
>sp|P81282|CSPG2_BOVIN Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP) Length = 3381 Score = 68.2 bits (165), Expect = 2e-11 Identities = 46/154 (29%), Positives = 72/154 (46%), Gaps = 6/154 (3%) Frame = +1 Query: 133 CLNGYTYIDGSWNDFPCTYKNLY--AICARDSLYIRFSFDYEVDSEKMIVSTDRYTYNEA 306 C NG T IDG +N F C Y A+C +D+ + + ++ + R T++ A Sbjct: 3121 CRNGATCIDG-FNTFRCLCLPSYVGALCEQDTETCDYGW-HKFQGQCYKYFAHRRTWDAA 3178 Query: 307 LQFCRTRGTQLARVNSASDNQAVASFAKRVVADYYWIDAND--YTEEGKWVDSNGKEKPY 480 + CR +G L + S + F RV DY WI ND + + +W D G Y Sbjct: 3179 ERECRLQGAHLTSILSHEEQM----FVNRVGHDYQWIGLNDKMFEHDFRWTD--GSTLQY 3232 Query: 481 KNWAAHEPNG--NRAENCVFSFYLEEGTWRDISC 576 +NW ++P+ + E+CV + E G W D+ C Sbjct: 3233 ENWRPNQPDSFFSTGEDCVVIIWHENGQWNDVPC 3266
Score = 39.7 bits (91), Expect = 0.008
Identities = 21/68 (30%), Positives = 33/68 (48%), Gaps = 4/68 (5%)
Frame = +1
Query: 10 DLYWLDGNDAKTEG--RWVDSNDNDLKHKGFHPGEPNG--GVNENCLNGYTYIDGSWNDF 177
D W+ ND E RW D + L+++ + P +P+ E+C+ + +G WND
Sbjct: 3207 DYQWIGLNDKMFEHDFRWTDGST--LQYENWRPNQPDSFFSTGEDCVVIIWHENGQWNDV 3264
Query: 178 PCTYKNLY 201
PC Y Y
Sbjct: 3265 PCNYHLTY 3272
>sp|O02659|MBL2_BOVIN Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) Length = 249 Score = 68.2 bits (165), Expect = 2e-11 Identities = 40/129 (31%), Positives = 64/129 (49%), Gaps = 1/129 (0%) Frame = +1 Query: 226 YIRFSFDYEVDSEKMIVSTDRYTYNEALQFCRTRGTQLARVNSASDNQAVASFAKRVVAD 405 ++ FS V + + + +NE C ++A +A +N+A+ K +V + Sbjct: 126 WLIFSLGKRVGKKAFFTNGKKMPFNEVKTLCAQFQGRVATPMNAEENRAL----KDLVTE 181 Query: 406 YYWIDANDYTEEGKWVDSNGKEKPYKNWAAHEP-NGNRAENCVFSFYLEEGTWRDISCYN 582 ++ D EGK+VD GK Y+NW EP N + E+CV L +GTW DI+C Sbjct: 182 EAFLGITDQETEGKFVDLTGKGVTYQNWNDGEPNNASPGEHCV--TLLSDGTWNDIACSA 239 Query: 583 KYLAICTFS 609 +L +C FS Sbjct: 240 SFLTVCEFS 248
Score = 38.9 bits (89), Expect = 0.014
Identities = 18/52 (34%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Frame = +1
Query: 34 DAKTEGRWVDSNDNDLKHKGFHPGEP-NGGVNENCLNGYTYIDGSWNDFPCT 186
D +TEG++VD + ++ ++ GEP N E+C+ DG+WND C+
Sbjct: 189 DQETEGKFVDLTGKGVTYQNWNDGEPNNASPGEHCVT--LLSDGTWNDIACS 238
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 85,461,073
Number of Sequences: 369166
Number of extensions: 1887762
Number of successful extensions: 5338
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4826
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5179
length of database: 68,354,980
effective HSP length: 107
effective length of database: 48,588,335
effective search space used: 6413660220
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail