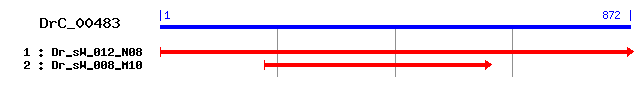
DrC_00483
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00483 (872 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q24307|IAP2_DROME Apoptosis 2 inhibitor (Inhibitor of ap... 66 1e-10 sp|P98170|BIRC4_HUMAN Baculoviral IAP repeat-containing pro... 65 2e-10 sp|Q86YT6|MIB1_HUMAN Ubiquitin ligase protein MIB1 (Mind bo... 64 4e-10 sp|Q13489|BIRC3_HUMAN Baculoviral IAP repeat-containing pro... 64 5e-10 sp|Q13490|BIRC2_HUMAN Baculoviral IAP repeat-containing pro... 62 2e-09 sp|Q6GNY1|MIB1_XENLA Ubiquitin ligase protein MIB1 (Mind bo... 62 2e-09 sp|Q80SY4|MIB1_MOUSE Ubiquitin ligase protein MIB1 (Mind bo... 62 2e-09 sp|O62640|PIAP_PIG Putative inhibitor of apoptosis 62 2e-09 sp|Q90660|BIR_CHICK Inhibitor of apoptosis protein (IAP) (I... 62 2e-09 sp|Q9R0I6|BIRC4_RAT Baculoviral IAP repeat-containing prote... 62 2e-09
>sp|Q24307|IAP2_DROME Apoptosis 2 inhibitor (Inhibitor of apoptosis 2) (dIAP2) (DIAP) (IAP homolog A) (IAP-like protein) (DILP) Length = 498 Score = 66.2 bits (160), Expect = 1e-10 Identities = 29/57 (50%), Positives = 41/57 (71%) Frame = +1 Query: 313 SVQEDNYQLEFIFPNNYCVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVCRTVIE 483 S++E+N QL+ C VCLD++V VVFLPCGHL TCN C+ +++CP+CR I+ Sbjct: 437 SLEEENRQLK---DARLCKVCLDEEVGVVFLPCGHLATCNQCAPSVANCPMCRADIK 490
>sp|P98170|BIRC4_HUMAN Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP) Length = 497 Score = 65.1 bits (157), Expect = 2e-10 Identities = 31/91 (34%), Positives = 50/91 (54%), Gaps = 12/91 (13%) Frame = +1 Query: 244 TGKDYDSTHEFLSDLKNYREIKHSVQEDNYQ------------LEFIFPNNYCVVCLDKQ 387 +G +Y S ++DL N + K S+Q+++ Q L + C +C+D+ Sbjct: 400 SGSNYKSLEVLVADLVNAQ--KDSMQDESSQTSLQKEISTEEQLRRLQEEKLCKICMDRN 457 Query: 388 VAVVFLPCGHLKTCNDCSNLLSSCPVCRTVI 480 +A+VF+PCGHL TC C+ + CP+C TVI Sbjct: 458 IAIVFVPCGHLVTCKQCAEAVDKCPMCYTVI 488
>sp|Q86YT6|MIB1_HUMAN Ubiquitin ligase protein MIB1 (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2) Length = 1006 Score = 64.3 bits (155), Expect = 4e-10 Identities = 25/43 (58%), Positives = 32/43 (74%) Frame = +1 Query: 364 CVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVCRTVIERRV 492 CVVC DK+ AV+F PCGH+ C +C+NL+ C CR V+ERRV Sbjct: 866 CVVCSDKKAAVLFQPCGHMCACENCANLMKKCVQCRAVVERRV 908
Score = 55.1 bits (131), Expect = 3e-07
Identities = 28/68 (41%), Positives = 39/68 (57%)
Frame = +1
Query: 289 KNYREIKHSVQEDNYQLEFIFPNNYCVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVC 468
K+ + VQ+ QL+ I C VCLD+ ++FL CGH TC C + +S CP+C
Sbjct: 938 KDNTNVNADVQKLQQQLQDIKEQTMCPVCLDRLKNMIFL-CGH-GTCQLCGDRMSECPIC 995
Query: 469 RTVIERRV 492
R IERR+
Sbjct: 996 RKAIERRI 1003
Score = 44.3 bits (103), Expect = 4e-04
Identities = 16/42 (38%), Positives = 26/42 (61%)
Frame = +1
Query: 364 CVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVCRTVIERR 489
C+VC D + +F PCGH+ TC+ CS + C +C+ ++ R
Sbjct: 819 CMVCSDMKRDTLFGPCGHIATCSLCSPRVKKCLICKEQVQSR 860
>sp|Q13489|BIRC3_HUMAN Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2) Length = 604 Score = 63.9 bits (154), Expect = 5e-10 Identities = 29/71 (40%), Positives = 42/71 (59%), Gaps = 6/71 (8%) Frame = +1 Query: 298 REIKHSVQED------NYQLEFIFPNNYCVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSC 459 ++IK+ ED QL + C VC+DK+V++VF+PCGHL C DC+ L C Sbjct: 529 QDIKYIPTEDVSDLPVEEQLRRLQEERTCKVCMDKEVSIVFIPCGHLVVCKDCAPSLRKC 588 Query: 460 PVCRTVIERRV 492 P+CR+ I+ V Sbjct: 589 PICRSTIKGTV 599
>sp|Q13490|BIRC2_HUMAN Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B) Length = 618 Score = 62.4 bits (150), Expect = 2e-09 Identities = 25/53 (47%), Positives = 35/53 (66%) Frame = +1 Query: 334 QLEFIFPNNYCVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVCRTVIERRV 492 QL + C VC+DK+V+VVF+PCGHL C +C+ L CP+CR +I+ V Sbjct: 561 QLRRLQEERTCKVCMDKEVSVVFIPCGHLVVCQECAPSLRKCPICRGIIKGTV 613
>sp|Q6GNY1|MIB1_XENLA Ubiquitin ligase protein MIB1 (Mind bomb homolog 1) Length = 1011 Score = 62.4 bits (150), Expect = 2e-09 Identities = 24/43 (55%), Positives = 32/43 (74%) Frame = +1 Query: 364 CVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVCRTVIERRV 492 CVVC DK+ AV+F PCGH+ C +C++L+ C CR V+ERRV Sbjct: 867 CVVCSDKKAAVLFQPCGHMCACENCASLMKKCVQCRAVVERRV 909
Score = 54.3 bits (129), Expect = 4e-07
Identities = 27/68 (39%), Positives = 39/68 (57%)
Frame = +1
Query: 289 KNYREIKHSVQEDNYQLEFIFPNNYCVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVC 468
K+ + VQ+ QL+ I C VCLD+ ++F+ CGH TC C + +S CP+C
Sbjct: 939 KDNTNVNADVQKLQQQLQDIKEQTMCPVCLDRLKNMIFM-CGH-GTCQLCGDRMSECPIC 996
Query: 469 RTVIERRV 492
R IERR+
Sbjct: 997 RKAIERRI 1004
Score = 43.1 bits (100), Expect = 0.001
Identities = 16/42 (38%), Positives = 26/42 (61%)
Frame = +1
Query: 364 CVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVCRTVIERR 489
C+VC D + +F PCGH+ TC+ CS + C +C+ ++ R
Sbjct: 820 CMVCSDLKRDTLFGPCGHIATCSLCSPRVKKCLLCKEQVQSR 861
>sp|Q80SY4|MIB1_MOUSE Ubiquitin ligase protein MIB1 (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) Length = 1006 Score = 62.4 bits (150), Expect = 2e-09 Identities = 24/43 (55%), Positives = 32/43 (74%) Frame = +1 Query: 364 CVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVCRTVIERRV 492 CVVC DK+ AV+F PCGH+ C +C++L+ C CR V+ERRV Sbjct: 866 CVVCSDKKAAVLFQPCGHMCACENCASLMKKCVQCRAVVERRV 908
Score = 55.1 bits (131), Expect = 3e-07
Identities = 28/68 (41%), Positives = 39/68 (57%)
Frame = +1
Query: 289 KNYREIKHSVQEDNYQLEFIFPNNYCVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVC 468
K+ + VQ+ QL+ I C VCLD+ ++FL CGH TC C + +S CP+C
Sbjct: 938 KDNTNVNADVQKLQQQLQDIKEQTMCPVCLDRLKNMIFL-CGH-GTCQLCGDRMSECPIC 995
Query: 469 RTVIERRV 492
R IERR+
Sbjct: 996 RKAIERRI 1003
Score = 44.3 bits (103), Expect = 4e-04
Identities = 16/42 (38%), Positives = 26/42 (61%)
Frame = +1
Query: 364 CVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVCRTVIERR 489
C+VC D + +F PCGH+ TC+ CS + C +C+ ++ R
Sbjct: 819 CMVCSDMKRDTLFGPCGHIATCSLCSPRVKKCLICKEQVQSR 860
>sp|O62640|PIAP_PIG Putative inhibitor of apoptosis Length = 358 Score = 62.4 bits (150), Expect = 2e-09 Identities = 25/53 (47%), Positives = 34/53 (64%) Frame = +1 Query: 334 QLEFIFPNNYCVVCLDKQVAVVFLPCGHLKTCNDCSNLLSSCPVCRTVIERRV 492 QL + C VC+DK+V++VF+PCGHL C DC+ L CP+CR I+ V Sbjct: 301 QLRRLQEERTCKVCMDKEVSIVFIPCGHLVVCKDCAPSLRKCPICRGTIKGTV 353
>sp|Q90660|BIR_CHICK Inhibitor of apoptosis protein (IAP) (Inhibitor of T cell apoptosis protein) Length = 611 Score = 62.4 bits (150), Expect = 2e-09 Identities = 32/87 (36%), Positives = 46/87 (52%), Gaps = 6/87 (6%) Frame = +1 Query: 250 KDYDSTHEFLSDLKNYREIKHSVQED------NYQLEFIFPNNYCVVCLDKQVAVVFLPC 411 KD+D DL + +K+ ED QL + C VC+DK+V++VF+PC Sbjct: 522 KDFDPV--LYKDLFVEKSMKYVPTEDVSGLPMEEQLRRLQEERTCKVCMDKEVSIVFIPC 579 Query: 412 GHLKTCNDCSNLLSSCPVCRTVIERRV 492 GHL C +C+ L CP+CR I+ V Sbjct: 580 GHLVVCKECAPSLRKCPICRGTIKGTV 606
>sp|Q9R0I6|BIRC4_RAT Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (RIAP3) (RIAP-3) Length = 496 Score = 62.0 bits (149), Expect = 2e-09 Identities = 30/89 (33%), Positives = 50/89 (56%), Gaps = 10/89 (11%) Frame = +1 Query: 244 TGKDYDSTHEFLSDLKNYRE-------IKHSVQED---NYQLEFIFPNNYCVVCLDKQVA 393 +G +Y S ++DL + ++ + S+Q+D QL + C +C+D+ +A Sbjct: 399 SGSNYLSLEVLIADLVSAQKDNSQDESSQTSLQKDISTEEQLRRLQEEKLCKICMDRNIA 458 Query: 394 VVFLPCGHLKTCNDCSNLLSSCPVCRTVI 480 +VF+PCGHL TC C+ + CP+C TVI Sbjct: 459 IVFVPCGHLVTCKQCAEAVDKCPMCCTVI 487
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 93,463,022
Number of Sequences: 369166
Number of extensions: 1890928
Number of successful extensions: 5402
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5181
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5372
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 8646143400
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail