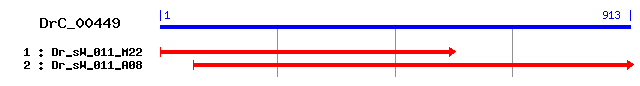
DrC_00449
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00449 (913 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9BUL8|PDC10_HUMAN Programmed cell death protein 10 (TF-... 114 4e-25 sp|Q6DF07|PDC10_XENTR Programmed cell death protein 10 114 4e-25 sp|Q8AVR4|PDC10_XENLA Programmed cell death protein 10 114 4e-25 sp|Q8VE70|PDC10_MOUSE Programmed cell death protein 10 (TF-... 114 5e-25 sp|Q6NX65|PDC10_RAT Programmed cell death protein 10 114 5e-25 sp|Q6PHH3|PDC10_BRARE Programmed cell death protein 10 113 6e-25 sp|Q5ZIV5|PDC10_CHICK Programmed cell death protein 10 112 1e-24 sp|P35875|PARP_DROME Poly [ADP-ribose] polymerase (PARP) (A... 36 0.17 sp|O42649|SMC3_SCHPO Structural maintenance of chromosome 3... 35 0.29 sp|P29461|PTP2_YEAST Tyrosine-protein phosphatase 2 (Protei... 33 0.85
>sp|Q9BUL8|PDC10_HUMAN Programmed cell death protein 10 (TF-1 cell apoptosis-related protein 15) (Cerebral cavernous malformations 3 protein) Length = 212 Score = 114 bits (285), Expect = 4e-25 Identities = 63/133 (47%), Positives = 88/133 (66%) Frame = +1 Query: 190 EVDMSETFLRMASMAETGVVRNMRKQIYNLLESKAHTLKEVLSRIPDEISDRRSFLDTIK 369 EV+ +E+ LRMA+ + + + L KA LK++LS+IPDEI+DR FL TIK Sbjct: 73 EVNFTESLLRMAADDVEEYMIERPEPEFQDLNEKARALKQILSKIPDEINDRVRFLQTIK 132 Query: 370 EIASAIKNLLDCVSEVITHLPTDGHKTSLEQAKRDFVKYSKRFSNTLKAFFRDNKPSEVY 549 +IASAIK LLD V+ V ++ +LE K++FVKYSK FS+TLK +F+D K V+ Sbjct: 133 DIASAIKELLDTVNNVFKKYQYQ-NRRALEHQKKEFVKYSKSFSDTLKTYFKDGKAINVF 191 Query: 550 ASANLLVNQTNMI 588 SAN L++QTN+I Sbjct: 192 VSANRLIHQTNLI 204
Score = 36.2 bits (82), Expect = 0.13
Identities = 16/37 (43%), Positives = 25/37 (67%)
Frame = +3
Query: 39 VFEPVFEKLEQKSLSHTHTLKQSFAKVEQQYPGFSQN 149
V PVF +LE+ +LS TL+ +F K E++ PG +Q+
Sbjct: 25 VMYPVFNELERVNLSAAQTLRAAFIKAEKENPGLTQD 61
>sp|Q6DF07|PDC10_XENTR Programmed cell death protein 10 Length = 212 Score = 114 bits (285), Expect = 4e-25 Identities = 63/133 (47%), Positives = 88/133 (66%) Frame = +1 Query: 190 EVDMSETFLRMASMAETGVVRNMRKQIYNLLESKAHTLKEVLSRIPDEISDRRSFLDTIK 369 EV+ +E+ LRMA+ + + + L KA LK++LS+IPDEI+DR FL TIK Sbjct: 73 EVNFTESLLRMAADDVEEYMVERPEPEFQELNEKARALKQILSKIPDEINDRVRFLQTIK 132 Query: 370 EIASAIKNLLDCVSEVITHLPTDGHKTSLEQAKRDFVKYSKRFSNTLKAFFRDNKPSEVY 549 +IASAIK LLD V+ V ++ +LE K++FVKYSK FS+TLK +F+D K V+ Sbjct: 133 DIASAIKELLDTVNNVFKKYQYQ-NRRALEHQKKEFVKYSKSFSDTLKTYFKDGKALNVF 191 Query: 550 ASANLLVNQTNMI 588 SAN L++QTN+I Sbjct: 192 ISANRLIHQTNLI 204
Score = 36.6 bits (83), Expect = 0.10
Identities = 17/45 (37%), Positives = 26/45 (57%)
Frame = +3
Query: 39 VFEPVFEKLEQKSLSHTHTLKQSFAKVEQQYPGFSQNFFNWYFEQ 173
V PVF +LE +LS TL+ +F K E++ PG +Q+ E+
Sbjct: 25 VMYPVFNELEHVNLSAAQTLRAAFIKAEKENPGLTQDIITKILEK 69
>sp|Q8AVR4|PDC10_XENLA Programmed cell death protein 10 Length = 212 Score = 114 bits (285), Expect = 4e-25 Identities = 63/133 (47%), Positives = 88/133 (66%) Frame = +1 Query: 190 EVDMSETFLRMASMAETGVVRNMRKQIYNLLESKAHTLKEVLSRIPDEISDRRSFLDTIK 369 EV+ +E+ LRMA+ + + + L KA LK++LS+IPDEI+DR FL TIK Sbjct: 73 EVNFTESLLRMAADDVEEYMVERPEPEFQELNEKARALKQILSKIPDEINDRVRFLQTIK 132 Query: 370 EIASAIKNLLDCVSEVITHLPTDGHKTSLEQAKRDFVKYSKRFSNTLKAFFRDNKPSEVY 549 +IASAIK LLD V+ V ++ +LE K++FVKYSK FS+TLK +F+D K V+ Sbjct: 133 DIASAIKELLDTVNNVFKKYQYQ-NRRALEHQKKEFVKYSKSFSDTLKTYFKDGKALNVF 191 Query: 550 ASANLLVNQTNMI 588 SAN L++QTN+I Sbjct: 192 ISANRLIHQTNLI 204
Score = 37.0 bits (84), Expect = 0.077
Identities = 17/45 (37%), Positives = 27/45 (60%)
Frame = +3
Query: 39 VFEPVFEKLEQKSLSHTHTLKQSFAKVEQQYPGFSQNFFNWYFEQ 173
V PVF +LE+ +LS TL+ +F K E++ PG +Q+ E+
Sbjct: 25 VMYPVFNELERVNLSAAQTLRAAFIKAEKENPGLTQDIITKILEK 69
>sp|Q8VE70|PDC10_MOUSE Programmed cell death protein 10 (TF-1 cell apoptosis-related protein 15) Length = 212 Score = 114 bits (284), Expect = 5e-25 Identities = 63/133 (47%), Positives = 88/133 (66%) Frame = +1 Query: 190 EVDMSETFLRMASMAETGVVRNMRKQIYNLLESKAHTLKEVLSRIPDEISDRRSFLDTIK 369 EV+ +E+ LRMA+ + + + L KA LK++LS+IPDEI+DR FL TIK Sbjct: 73 EVNFTESLLRMAADDVEEYMIERPEPEFQDLNEKARALKQILSKIPDEINDRVRFLQTIK 132 Query: 370 EIASAIKNLLDCVSEVITHLPTDGHKTSLEQAKRDFVKYSKRFSNTLKAFFRDNKPSEVY 549 +IASAIK LLD V+ V ++ +LE K++FVKYSK FS+TLK +F+D K V+ Sbjct: 133 DIASAIKELLDTVNNVFKKYQYQ-NRRALEHQKKEFVKYSKSFSDTLKTYFKDGKAINVF 191 Query: 550 ASANLLVNQTNMI 588 SAN L++QTN+I Sbjct: 192 ISANRLIHQTNLI 204
Score = 36.2 bits (82), Expect = 0.13
Identities = 16/37 (43%), Positives = 25/37 (67%)
Frame = +3
Query: 39 VFEPVFEKLEQKSLSHTHTLKQSFAKVEQQYPGFSQN 149
V PVF +LE+ +LS TL+ +F K E++ PG +Q+
Sbjct: 25 VMYPVFNELERVNLSAAQTLRAAFIKAEKENPGLTQD 61
>sp|Q6NX65|PDC10_RAT Programmed cell death protein 10 Length = 210 Score = 114 bits (284), Expect = 5e-25 Identities = 63/133 (47%), Positives = 88/133 (66%) Frame = +1 Query: 190 EVDMSETFLRMASMAETGVVRNMRKQIYNLLESKAHTLKEVLSRIPDEISDRRSFLDTIK 369 EV+ +E+ LRMA+ + + + L KA LK++LS+IPDEI+DR FL TIK Sbjct: 71 EVNFTESLLRMAADDVEEYMIERPEPEFQDLNEKARALKQILSKIPDEINDRVRFLQTIK 130 Query: 370 EIASAIKNLLDCVSEVITHLPTDGHKTSLEQAKRDFVKYSKRFSNTLKAFFRDNKPSEVY 549 +IASAIK LLD V+ V ++ +LE K++FVKYSK FS+TLK +F+D K V+ Sbjct: 131 DIASAIKELLDTVNNVFKKYQYQ-NRRALEHQKKEFVKYSKSFSDTLKTYFKDGKAINVF 189 Query: 550 ASANLLVNQTNMI 588 SAN L++QTN+I Sbjct: 190 ISANRLIHQTNLI 202
Score = 36.2 bits (82), Expect = 0.13
Identities = 16/37 (43%), Positives = 25/37 (67%)
Frame = +3
Query: 39 VFEPVFEKLEQKSLSHTHTLKQSFAKVEQQYPGFSQN 149
V PVF +LE+ +LS TL+ +F K E++ PG +Q+
Sbjct: 23 VMYPVFNELERVNLSAAQTLRAAFIKAEKENPGLTQD 59
>sp|Q6PHH3|PDC10_BRARE Programmed cell death protein 10 Length = 205 Score = 113 bits (283), Expect = 6e-25 Identities = 61/133 (45%), Positives = 90/133 (67%) Frame = +1 Query: 190 EVDMSETFLRMASMAETGVVRNMRKQIYNLLESKAHTLKEVLSRIPDEISDRRSFLDTIK 369 E++ +E+ LRMA+ + + ++ + L +A LK++LS+IPDEI+DR FL TIK Sbjct: 66 EINFTESLLRMAADDVEEYMIDRPEREFQDLNERARALKQILSKIPDEINDRVRFLQTIK 125 Query: 370 EIASAIKNLLDCVSEVITHLPTDGHKTSLEQAKRDFVKYSKRFSNTLKAFFRDNKPSEVY 549 +IASAIK LLD V+ V + +LE K++FVK+SK FS+TLK +F+D K V+ Sbjct: 126 DIASAIKELLDTVNNVFKKYQYQNRR-ALEHQKKEFVKHSKSFSDTLKTYFKDGKAINVF 184 Query: 550 ASANLLVNQTNMI 588 ASAN L++QTN+I Sbjct: 185 ASANRLIHQTNLI 197
Score = 35.8 bits (81), Expect = 0.17
Identities = 16/37 (43%), Positives = 24/37 (64%)
Frame = +3
Query: 39 VFEPVFEKLEQKSLSHTHTLKQSFAKVEQQYPGFSQN 149
V PVF +LE +LS TL+ +F K E++ PG +Q+
Sbjct: 18 VMYPVFNELESVNLSAAQTLRAAFKKAEKENPGLTQD 54
>sp|Q5ZIV5|PDC10_CHICK Programmed cell death protein 10 Length = 212 Score = 112 bits (281), Expect = 1e-24 Identities = 62/133 (46%), Positives = 88/133 (66%) Frame = +1 Query: 190 EVDMSETFLRMASMAETGVVRNMRKQIYNLLESKAHTLKEVLSRIPDEISDRRSFLDTIK 369 +V+ +E+ LRMA+ + + + L KA LK++LS+IPDEI+DR FL TIK Sbjct: 73 DVNFTESLLRMAADDVEEYMIERPEPEFQDLNEKARALKQILSKIPDEINDRVRFLQTIK 132 Query: 370 EIASAIKNLLDCVSEVITHLPTDGHKTSLEQAKRDFVKYSKRFSNTLKAFFRDNKPSEVY 549 +IASAIK LLD V+ V ++ +LE K++FVKYSK FS+TLK +F+D K V+ Sbjct: 133 DIASAIKELLDTVNNVFKKYQYQ-NRRALEHQKKEFVKYSKSFSDTLKTYFKDGKAINVF 191 Query: 550 ASANLLVNQTNMI 588 SAN L++QTN+I Sbjct: 192 ISANRLIHQTNLI 204
Score = 36.2 bits (82), Expect = 0.13
Identities = 16/37 (43%), Positives = 25/37 (67%)
Frame = +3
Query: 39 VFEPVFEKLEQKSLSHTHTLKQSFAKVEQQYPGFSQN 149
V PVF +LE+ +LS TL+ +F K E++ PG +Q+
Sbjct: 25 VMYPVFNELERVNLSAAQTLRAAFIKAEKENPGLTQD 61
>sp|P35875|PARP_DROME Poly [ADP-ribose] polymerase (PARP) (ADPRT) (NAD(+) ADP-ribosyltransferase) (Poly[ADP-ribose] synthetase) Length = 994 Score = 35.8 bits (81), Expect = 0.17 Identities = 38/147 (25%), Positives = 62/147 (42%), Gaps = 22/147 (14%) Frame = +1 Query: 127 NIQDFLK-IFLIGILNKLGLENEVDMSETFLRMASMAETGVVRNMRKQIYNLLESKAHTL 303 ++Q+ +K IF I +NK +E +DM + L S + + K+IYN+LE ++T Sbjct: 650 SVQNLIKLIFDIDSMNKTLMEFHIDMDKMPLGKLSAHQIQSAYRVVKEIYNVLECGSNTA 709 Query: 304 KEV--------------------LSRIPDEISDRRSFLDTIKEIASAIKNL-LDCVSEVI 420 K + L +I D R LD++ EI A + + VS+ Sbjct: 710 KLIDATNRFYTLIPHNFGVQLPTLIETHQQIEDLRQMLDSLAEIEVAYSIIKSEDVSDAC 769 Query: 421 THLPTDGHKTSLEQAKRDFVKYSKRFS 501 P D H ++ K S+ FS Sbjct: 770 N--PLDNHYAQIKTQLVALDKNSEEFS 794
>sp|O42649|SMC3_SCHPO Structural maintenance of chromosome 3 (Cohesin complex Psm3 subunit) Length = 1194 Score = 35.0 bits (79), Expect = 0.29 Identities = 30/113 (26%), Positives = 51/113 (45%), Gaps = 2/113 (1%) Frame = +1 Query: 172 KLGLENEVDMSETFLRMASMAETGVVRNMRKQIYNL--LESKAHTLKEVLSRIPDEISDR 345 ++G +N +D SE V R++ K L ++S + L+E + RI EISD+ Sbjct: 835 EIGSDNRIDESEL---------NSVKRSLLKYENKLQIIKSSSSGLEEQMQRINSEISDK 885 Query: 346 RSFLDTIKEIASAIKNLLDCVSEVITHLPTDGHKTSLEQAKRDFVKYSKRFSN 504 R+ L++++E+ EV T + D AKR + K+ N Sbjct: 886 RNELESLEELQ----------HEVATRIEQDAKINERNAAKRSLLLARKKECN 928
>sp|P29461|PTP2_YEAST Tyrosine-protein phosphatase 2 (Protein-tyrosine phosphatase 2) (PTPase 2) Length = 750 Score = 33.5 bits (75), Expect = 0.85 Identities = 16/48 (33%), Positives = 28/48 (58%) Frame = +1 Query: 430 PTDGHKTSLEQAKRDFVKYSKRFSNTLKAFFRDNKPSEVYASANLLVN 573 P D +K L+ K+D + YS N+L+ FF+ N P+++ + +L N Sbjct: 239 PNDSNKIFLQSLKKDLIHYS---PNSLQKFFQFNMPADLAPNDTILPN 283
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 103,679,187
Number of Sequences: 369166
Number of extensions: 2129695
Number of successful extensions: 5552
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5294
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5529
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 9270587090
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail