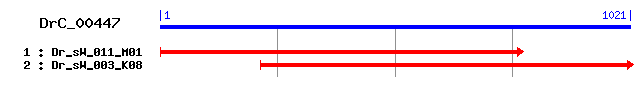
DrC_00447
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00447 (1021 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O17453|PSD4_SCHMA 26S proteasome non-ATPase regulatory s... 256 6e-68 sp|P55036|PSD4_HUMAN 26S proteasome non-ATPase regulatory s... 238 2e-62 sp|O35226|PSD4_MOUSE 26S proteasome non-ATPase regulatory s... 236 6e-62 sp|P55035|PSD4_DROME 26S proteasome non-ATPase regulatory s... 209 1e-53 sp|P55034|PSD4_ARATH 26S proteasome non-ATPase regulatory s... 158 2e-38 sp|P38886|RPN10_YEAST 26S proteasome regulatory subunit RPN10 142 2e-33 sp|O94444|RPN10_SCHPO 26S proteasome regulatory subunit rpn10 120 8e-27 sp|Q6GDD5|ICAC_STAAR Biofilm PIA synthesis protein icaC (In... 31 5.0
>sp|O17453|PSD4_SCHMA 26S proteasome non-ATPase regulatory subunit 4 (26S proteasome regulatory subunit S5A) Length = 420 Score = 256 bits (655), Expect = 6e-68 Identities = 142/323 (43%), Positives = 197/323 (60%), Gaps = 18/323 (5%) Frame = +1 Query: 1 DFMRNGDYNPNRLQAQIDAFNYFTNAKKRMNVENTCGLVTLANYQVVCTLTTDMSKLHSK 180 D+MRNGD+ P RLQAQ DA +K++ N ENT GL++LAN +V+CTLT D+SK++++ Sbjct: 14 DYMRNGDFFPTRLQAQNDAVGLICQSKRQRNPENTIGLLSLANTEVLCTLTNDVSKIYNR 73 Query: 181 ITLVDLSGNINFCTTVRVSQLALRHRQSRNQKPRIVVFVGSPIYXXXXXXXXXXXXXXXX 360 + LV+ G I FC+++R++ LALRHRQ R+QK RIV F+GSPI Sbjct: 74 LHLVEPKGRIIFCSSIRIAHLALRHRQLRHQKMRIVCFIGSPILEDEKELTRLAKRLKKE 133 Query: 361 XVNIDVISFGESEENQHKLSAFIDAINGKDGSGSHLVTIPAGSSLQESLRTSPILMNEES 540 VN+D+I+FGE+E N+ KLS FID +NGKDG+GSHL+++ G+ L ++L TSP++ E+ Sbjct: 134 KVNVDIINFGENETNEQKLSEFIDTLNGKDGTGSHLISVAPGTVLHDTLMTSPVVAGEDG 193 Query: 541 GSLGAVG----FGMEEIDDPDLLYALRVSMEDQRMQQERA----XXXXXXXXXXXXXXXX 696 + G FG++ +DPDLLYALRVSMEDQRM+QE Sbjct: 194 SGMAGAGLGLEFGLDGAEDPDLLYALRVSMEDQRMRQEHEVNGDGSNTSVVATSLPAGSG 253 Query: 697 XSEEDMLREAMAASLSDNIGE-------VDFAAMTEEEQIAYATRMSLQ---DEAGAVGS 846 SEE ML++A+A S+ N E +D AAM+EE+QIAYA RMSLQ +E + Sbjct: 254 TSEEAMLQQALAMSMQMNNTESSSLPMDIDLAAMSEEDQIAYALRMSLQQMGEETTQPTT 313 Query: 847 FGETMDTVVEEPEKEQFDVVDNP 915 D + EP D+ P Sbjct: 314 TTLESDKTIVEPSGVAMDIDQTP 336
>sp|P55036|PSD4_HUMAN 26S proteasome non-ATPase regulatory subunit 4 (26S proteasome regulatory subunit S5A) (Rpn10) (Multiubiquitin chain binding protein) (Antisecretory factor 1) (AF) (ASF) Length = 377 Score = 238 bits (608), Expect = 2e-62 Identities = 140/329 (42%), Positives = 192/329 (58%), Gaps = 14/329 (4%) Frame = +1 Query: 1 DFMRNGDYNPNRLQAQIDAFNYFTNAKKRMNVENTCGLVTLAN-YQVVCTLTTDMSKLHS 177 ++MRNGD+ P RLQAQ DA N ++K R N EN GL+TLAN +V+ TLT D ++ S Sbjct: 14 EYMRNGDFLPTRLQAQQDAVNIVCHSKTRSNPENNVGLITLANDCEVLTTLTPDTGRILS 73 Query: 178 KITLVDLSGNINFCTTVRVSQLALRHRQSRNQKPRIVVFVGSPIYXXXXXXXXXXXXXXX 357 K+ V G I FCT +RV+ LAL+HRQ +N K RI+ FVGSP+ Sbjct: 74 KLHTVQPKGKITFCTGIRVAHLALKHRQGKNHKMRIIAFVGSPVEDNEKDLVKLAKRLKK 133 Query: 358 XXVNIDVISFGESEENQHKLSAFIDAINGKDGSGSHLVTIPAGSSLQESLRTSPILMNEE 537 VN+D+I+FGE E N KL+AF++ +NGKDG+GSHLVT+P G SL ++L +SPIL E Sbjct: 134 EKVNVDIINFGEEEVNTEKLTAFVNTLNGKDGTGSHLVTVPPGPSLADALISSPILAGEG 193 Query: 538 SGSLGA----VGFGMEEIDDPDLLYALRVSMEDQRMQQE----RAXXXXXXXXXXXXXXX 693 LG FG++ DP+L ALRVSME+QR +QE RA Sbjct: 194 GAMLGLGASDFEFGVDPSADPELALALRVSMEEQRQRQEEEARRAAAASAAEAGIATTGT 253 Query: 694 XXSEEDMLREAMAASLSDNIGEVDFAAMTEEEQIAYATRMSLQD-EAGAVGS----FGET 858 S++ +L+ ++ G D ++MTEEEQIAYA +MSLQ E G S Sbjct: 254 EDSDDALLKMTISQQEFGRTGLPDLSSMTEEEQIAYAMQMSLQGAEFGQAESADIDASSA 313 Query: 859 MDTVVEEPEKEQFDVVDNPDLIGAVLEGM 945 MDT E++ +DV+ +P+ + +VLE + Sbjct: 314 MDTSEPAKEEDDYDVMQDPEFLQSVLENL 342
>sp|O35226|PSD4_MOUSE 26S proteasome non-ATPase regulatory subunit 4 (26S proteasome regulatory subunit S5A) (Rpn10) (Multiubiquitin chain binding protein) Length = 376 Score = 236 bits (603), Expect = 6e-62 Identities = 138/328 (42%), Positives = 190/328 (57%), Gaps = 13/328 (3%) Frame = +1 Query: 1 DFMRNGDYNPNRLQAQIDAFNYFTNAKKRMNVENTCGLVTLAN-YQVVCTLTTDMSKLHS 177 ++MRNGD+ P RLQAQ DA N ++K R N EN GL+TLAN +V+ TLT D ++ S Sbjct: 14 EYMRNGDFLPTRLQAQQDAVNIVCHSKTRSNPENNVGLITLANDCEVLTTLTPDTGRILS 73 Query: 178 KITLVDLSGNINFCTTVRVSQLALRHRQSRNQKPRIVVFVGSPIYXXXXXXXXXXXXXXX 357 K+ V G I FCT +RV+ LAL+HRQ +N K RI+ FVGSP+ Sbjct: 74 KLHTVQPKGKITFCTGIRVAHLALKHRQGKNHKMRIIAFVGSPVEDNEKDLVKLAKRLKK 133 Query: 358 XXVNIDVISFGESEENQHKLSAFIDAINGKDGSGSHLVTIPAGSSLQESLRTSPILMNEE 537 VN+D+I+FGE E N KL+AF++ +NGKDG+GSHLVT+P G SL ++L +SPIL E Sbjct: 134 EKVNVDIINFGEEEVNTEKLTAFVNTLNGKDGTGSHLVTVPPGPSLADALISSPILAGEG 193 Query: 538 SGSLGA----VGFGMEEIDDPDLLYALRVSMEDQRMQQE----RAXXXXXXXXXXXXXXX 693 LG FG++ DP+L ALRVSME+QR +QE RA Sbjct: 194 GAMLGLGASDFEFGVDPSADPELALALRVSMEEQRQRQEEEARRAAAASAAEAGIATPGT 253 Query: 694 XXSEEDMLREAMAASLSDNIGEVDFAAMTEEEQIAYATRMSLQ----DEAGAVGSFGETM 861 S++ +L+ + G D ++MTEEEQIAYA +MSLQ + A M Sbjct: 254 EDSDDALLKMTINQQEFGRPGLPDLSSMTEEEQIAYAMQMSLQGTEFSQESADMDASSAM 313 Query: 862 DTVVEEPEKEQFDVVDNPDLIGAVLEGM 945 DT E++ +DV+ +P+ + +VLE + Sbjct: 314 DTSDPVKEEDDYDVMQDPEFLQSVLENL 341
>sp|P55035|PSD4_DROME 26S proteasome non-ATPase regulatory subunit 4 (26S proteasome regulatory subunit S5A) (Multiubiquitin chain binding protein) (54 kDa subunit of mu particle) (p54) Length = 396 Score = 209 bits (531), Expect = 1e-53 Identities = 133/337 (39%), Positives = 182/337 (54%), Gaps = 32/337 (9%) Frame = +1 Query: 1 DFMRNGDYNPNRLQAQIDAFNYFTNAKKRMNVENTCGLVTLAN-YQVVCTLTTDMSKLHS 177 DF RNGDY P RL Q D N K R N EN GL+TL+N +V+ TLT+D ++ S Sbjct: 14 DFQRNGDYFPTRLIVQRDGINLVCLTKLRSNPENNVGLMTLSNTVEVLATLTSDAGRIFS 73 Query: 178 KITLVDLSGNINFCTTVRVSQLALRHRQSRNQKPRIVVFVGSPIYXXXXXXXXXXXXXXX 357 K+ LV G IN T +R++ L L+HRQ +N K RIVVFVGSPI Sbjct: 74 KMHLVQPKGEINLLTGIRIAHLVLKHRQGKNHKMRIVVFVGSPINHEEGDLVKQAKRLKK 133 Query: 358 XXVNIDVISFGESEENQHKLSAFIDAINGKDGSGSHLVTIPAGSSLQESLRTSPILMNEE 537 VN+D++SFG+ N L+AFI+A+NGKDG+GSHLV++P GS L ++L +SPI+ E+ Sbjct: 134 EKVNVDIVSFGDHGNNNEILTAFINALNGKDGTGSHLVSVPRGSVLSDALLSSPIIQGED 193 Query: 538 S-GSLGAVG----FGMEEIDDPDLLYALRVSMEDQRMQQER------------------- 645 G G G FG++ +DP+L ALRVSME+QR +QE Sbjct: 194 GMGGAGLGGNVFEFGVDPNEDPELALALRVSMEEQRQRQESEQRRANPDGAPPTGGDAGG 253 Query: 646 ----AXXXXXXXXXXXXXXXXXSEEDMLREAMAASL---SDNIGEVDFAAMTEEEQIAYA 804 + +EE ML+ A+A S DN+ DFA MTEEEQIA+A Sbjct: 254 GGGVSGSGPGNEESAGAENEANTEEAMLQRALALSTETPEDNL--PDFANMTEEEQIAFA 311 Query: 805 TRMSLQDEAGAVGSFGETMDTVVEEPEKEQFDVVDNP 915 +MS+QD D+V ++ ++ + D + P Sbjct: 312 MQMSMQDAPD---------DSVTQQAKRPKTDEANAP 339
>sp|P55034|PSD4_ARATH 26S proteasome non-ATPase regulatory subunit 4 (26S proteasome regulatory subunit S5A) (Multiubiquitin chain binding protein) Length = 386 Score = 158 bits (400), Expect = 2e-38 Identities = 102/310 (32%), Positives = 168/310 (54%), Gaps = 34/310 (10%) Frame = +1 Query: 1 DFMRNGDYNPNRLQAQIDAFNYFTNAKKRMNVENTCGLVTLAN--YQVVCTLTTDMSKLH 174 ++MRNGDY+P+RLQAQ +A N AK + N ENT G++T+A +V+ T T+D+ K+ Sbjct: 14 EWMRNGDYSPSRLQAQTEAVNLLCGAKTQSNPENTVGILTMAGKGVRVLTTPTSDLGKIL 73 Query: 175 SKITLVDLSGNINFCTTVRVSQLALRHRQSRNQKPRIVVFVGSPIYXXXXXXXXXXXXXX 354 + + +D+ G IN ++++QLAL+HRQ++NQ+ RI+VF GSPI Sbjct: 74 ACMHGLDVGGEINLTAAIQIAQLALKHRQNKNQRQRIIVFAGSPIKYEKKALEIVGKRLK 133 Query: 355 XXXVNIDVISFGE--SEENQHKLSAFIDAINGKDGSGSHLVTIPAG-SSLQESLRTSPIL 525 V++D+++FGE EE KL A + A+N D GSH+V +P+G ++L + L ++P+ Sbjct: 134 KNSVSLDIVNFGEDDDEEKPQKLEALLTAVNNND--GSHIVHVPSGANALSDVLLSTPVF 191 Query: 526 MNEESGS------------LGAVGFGMEEIDDPDLLYALRVSMEDQRMQQERA---XXXX 660 +E S G FG++ DP+L ALRVSME++R +QE A Sbjct: 192 TGDEGASGYVSAAAAAAAAGGDFDFGVDPNIDPELALALRVSMEEERARQEAAAKKAADE 251 Query: 661 XXXXXXXXXXXXXSEEDMLREA-------------MAASLSDNIGEVDFAAMTEEEQ-IA 798 S+E + R + +++ ++G+V+ + +E+Q +A Sbjct: 252 AGQKDKDGDTASASQETVARTTDKNAEPMDEDSALLDQAIAMSVGDVNMSEAADEDQDLA 311 Query: 799 YATRMSLQDE 828 A +MS+ E Sbjct: 312 LALQMSMSGE 321
>sp|P38886|RPN10_YEAST 26S proteasome regulatory subunit RPN10 Length = 268 Score = 142 bits (358), Expect = 2e-33 Identities = 82/232 (35%), Positives = 128/232 (55%), Gaps = 17/232 (7%) Frame = +1 Query: 1 DFMRNGDYNPNRLQAQIDAFNYFTNAKKRMNVENTCGLVTLA--NYQVVCTLTTDMSKLH 174 ++ RNGD+ R +AQID+ + AK+ N ENT GL++ A N +V+ T T + K+ Sbjct: 14 EYSRNGDFPRTRFEAQIDSVEFIFQAKRNSNPENTVGLISGAGANPRVLSTFTAEFGKIL 73 Query: 175 SKITLVDLSGNINFCTTVRVSQLALRHRQSRNQKPRIVVFVGSPIYXXXXXXXXXXXXXX 354 + + + G ++ T ++++QL L+HRQ++ Q RIV FV SPI Sbjct: 74 AGLHDTQIEGKLHMATALQIAQLTLKHRQNKVQHQRIVAFVCSPISDSRDELIRLAKTLK 133 Query: 355 XXXVNIDVISFGESEENQHKLSAFIDAINGKDGSGSHLVTI-PAGSSLQESLRTSPILMN 531 V +D+I+FGE E+N L FI A+N SHL+T+ P L E++ +SPI++ Sbjct: 134 KNNVAVDIINFGEIEQNTELLDEFIAAVNNPQEETSHLLTVTPGPRLLYENIASSPIILE 193 Query: 532 EESGSLGAVG--------------FGMEEIDDPDLLYALRVSMEDQRMQQER 645 E S +GA G FG++ DP+L ALR+SME+++ +QER Sbjct: 194 EGSSGMGAFGGSGGDSDANGTFMDFGVDPSMDPELAMALRLSMEEEQQRQER 245
>sp|O94444|RPN10_SCHPO 26S proteasome regulatory subunit rpn10 Length = 243 Score = 120 bits (300), Expect = 8e-27 Identities = 79/218 (36%), Positives = 112/218 (51%), Gaps = 2/218 (0%) Frame = +1 Query: 1 DFMRNGDYNPNRLQAQIDAFNYFTNAKKRMNVENTCGLVTLANY--QVVCTLTTDMSKLH 174 ++M NGDY P R +AQ D + N K N EN CGL+T+ + QV+ TLT D K Sbjct: 14 EWMINGDYIPTRFEAQKDTVHMIFNQKINDNPENMCGLMTIGDNSPQVLSTLTRDYGKFL 73 Query: 175 SKITLVDLSGNINFCTTVRVSQLALRHRQSRNQKPRIVVFVGSPIYXXXXXXXXXXXXXX 354 S + + + GN F ++++QLAL+HR+++ Q+ RIV FVGSPI Sbjct: 74 SAMHDLPVRGNAKFGDGIQIAQLALKHRENKIQRQRIVAFVGSPIVEDEKNLIRLAKRMK 133 Query: 355 XXXVNIDVISFGESEENQHKLSAFIDAINGKDGSGSHLVTIPAGSSLQESLRTSPILMNE 534 V ID+I GE +N+ L FIDA N D HLV+IP L L + Sbjct: 134 KNNVAIDIIHIGEL-QNESALQHFIDAANSSD--SCHLVSIPPSPQLLSDLVNQSPIGQG 190 Query: 535 ESGSLGAVGFGMEEIDDPDLLYALRVSMEDQRMQQERA 648 S +G++ D +L AL +SM ++R +QE A Sbjct: 191 VVASQNQFEYGVDPNLDVELALALELSMAEERARQEVA 228
>sp|Q6GDD5|ICAC_STAAR Biofilm PIA synthesis protein icaC (Intercellular adhesion protein C) Length = 350 Score = 31.2 bits (69), Expect = 5.0 Identities = 18/71 (25%), Positives = 36/71 (50%), Gaps = 7/71 (9%) Frame = -2 Query: 1014 LKLYSNNLIVFSTIRFIIIDIMIH-------TFKYCTY*IRIIHDIKLFFFRFFNHSIHC 856 L+ Y N+++F T FII+ ++ T++Y T ++ I + F+++S Sbjct: 42 LQFYIRNIVIFGTPCFIILSQLLTTLNYQKVTYRYLTTRVKYILIPYILMGLFYSYSESL 101 Query: 855 LTETTNSSSFV 823 LT+T+ F+ Sbjct: 102 LTDTSFQKQFI 112
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 99,138,876
Number of Sequences: 369166
Number of extensions: 1816836
Number of successful extensions: 4900
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 4745
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4872
length of database: 68,354,980
effective HSP length: 111
effective length of database: 47,849,395
effective search space used: 10909662060
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail