DrC_00333
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00333 (1358 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q15036|SNX17_HUMAN Sorting nexin-17 183 9e-46 sp|Q19532|YTDK_CAEEL Hypothetical protein F17H10.3 in chrom... 94 1e-18 sp|Q7NAK6|GIDA_MYCGA tRNA uridine 5-carboxymethylaminomethy... 33 2.5 sp|Q9Z6S3|TRMB_CHLPN tRNA (guanine-N(7)-)-methyltransferase... 32 3.3 sp|Q92J94|RS7_RICCN 30S ribosomal protein S7 32 5.6 sp|P54939|TLN1_CHICK Talin-1 31 7.3 sp|P25386|USO1_YEAST Intracellular protein transport protei... 31 7.3 sp|Q9H2U9|ADAM7_HUMAN ADAM 7 precursor (A disintegrin and m... 31 7.3 sp|Q28475|ADAM7_MACFA ADAM 7 precursor (A disintegrin and m... 31 9.5 sp|Q92G13|HEMK_RICCN Bifunctional methyltransferase [Includ... 31 9.5
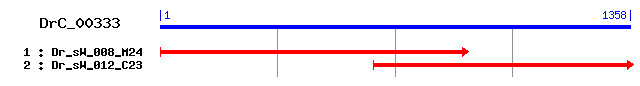
>sp|Q15036|SNX17_HUMAN Sorting nexin-17 Length = 470 Score = 183 bits (465), Expect = 9e-46 Identities = 117/374 (31%), Positives = 203/374 (54%), Gaps = 3/374 (0%) Frame = +3 Query: 3 YINGAHHCSARFSRLYKLYEDLKLRFPVSDLPYFPPKKFFKINXXXXXXXXXXXXXXXXK 182 ++NG HC R+S+L L+E L+ + + LP FPPKK F + Sbjct: 26 HVNGVLHCRVRYSQLLGLHEQLRKEYGANVLPAFPPKKLFSLTPAEVEQRREQLEKYMQA 85 Query: 183 VSSSNKFNSDE-LHDFLKTCQKETKKIVSEPVKLSVYLMDGQKHELDVWSTDNTKTVFEA 359 V S E + FL+ Q+ET+++ +E V L V L +GQK ++V ++D T+ V EA Sbjct: 86 VRQDPLLGSSETFNSFLRRAQQETQQVPTEEVSLEVLLSNGQKVLVNVLTSDQTEDVLEA 145 Query: 360 FADVISLPRKISSKFSLFFMVVHPDMKFSIIRKLQDFEYPYVSLYEIQKLCNCRIVVRKS 539 A + LP + FSLF + D FS +RKLQ+FE PYVS+ ++ +IV+RKS Sbjct: 146 VAAKLDLPDDLIGYFSLFLVREKEDGAFSFVRKLQEFELPYVSVTSLRSQ-EYKIVLRKS 204 Query: 540 VWNIELESEIDSHNSGLEILFAQVSAEIVNGWISIPDEARMNRIQNLIERKARKETLQLA 719 W+ + ++ + GL +L+AQ ++I GWI + E + ++++L E+ ++KE L+LA Sbjct: 205 YWDSAYDDDVMENRVGLNLLYAQTVSDIERGWILVTKE-QHRQLKSLQEKVSKKEFLRLA 263 Query: 720 RTLSYYGYMQLHGTSVCNRPLPNTMAIVSTGNFEIVFRDEHDDHVLMK--YYLTRIKAWK 893 +TL +YGY++ V + P + +VS GN E+ + L + + +TR++ W+ Sbjct: 264 QTLRHYGYLRF-DACVADFPEKDCPVVVSAGNSELSLQLRLPGQQLREGSFRVTRMRCWR 322 Query: 894 ISTALKEPLNDDINQSVNQSQALSPTLTFEYLDIETKKLDEVVISNNQTDQIILLSMSLK 1073 +++++ P + + + + L FEYL + +L V I+ + Q I++S+ L+ Sbjct: 323 VTSSVPLPSGSTSSPGRGRGE-VRLELAFEYL-MSKDRLQWVTIT---SPQAIMMSICLQ 377 Query: 1074 NMAEEYVCYKSSNS 1115 +M +E + KS S Sbjct: 378 SMVDELMVKKSGGS 391
>sp|Q19532|YTDK_CAEEL Hypothetical protein F17H10.3 in chromosome X Length = 540 Score = 93.6 bits (231), Expect = 1e-18 Identities = 103/435 (23%), Positives = 184/435 (42%), Gaps = 27/435 (6%) Frame = +3 Query: 3 YINGAHHCSARFSRLYKLYEDLKLRFPVSDL-PYFPPKKFFKINXXXXXXXXXXXXXXXX 179 +ING +H S RFS LY+ E +K +F P FP KK FK++ Sbjct: 92 HINGWYHGSVRFSHLYEFAELIKQKFSQRYKGPEFPAKKLFKLDPKAIDERRQKITKYFQ 151 Query: 180 KVSSSNKFNSDELHDF-LKTCQKETKKIVSEPVKLSVYLMDGQKHELDVWSTDNTKTVFE 356 + + L + L Q ++ + S+ V L V+L +G+K + +D+T + + Sbjct: 152 ALVQHPEVARHYLVEKKLLGFQIDSFRATSQYVTLDVFLGNGEKTTIKCLVSDSTLEIMK 211 Query: 357 AFADVISLPRK--ISSKFSLF-----------FMVVHPDMKFSIIRKLQDFEYPYVSLYE 497 + + K F LF + V + + R L++FE P+VSL Sbjct: 212 IICEKLGFKNKDQFIYHFGLFMAKGRDPTNACYSVTTENFNPLLTRFLRNFEAPFVSLST 271 Query: 498 IQKLCNCR-----IVVRKSVWNIELESEIDSHNSGLEILFAQVSAEIVNGWISIPDEARM 662 + N + +RK +W+ +E + + +E+L+ Q + NG + E Sbjct: 272 ANQKYNENGHYHFLCLRKLIWDSRVEEPLLDDGNFVELLYKQAMQDYKNGHMDPVKEDLD 331 Query: 663 NRIQNLIERKARKETLQLARTLSYYGYMQLHGTSVCNRPLPNTMAIVSTGNFEIVF--RD 836 +++++ + R K L+ LS Y Y ++ C+ P P T + G +I+ RD Sbjct: 332 SKLKSCMARNDSKMFLRTCHQLSTYSY-EIMSPCSCDYPKPGTPCEIKFGRRQIIMTTRD 390 Query: 837 EHDDHVLMKYYLTRIKAWKISTALKEPLNDDINQSVNQSQALSPTLTFEYLDIETKKLDE 1016 E + TRI+ W+I+ + D I+ Q + L TFE++ ++ Sbjct: 391 ETGKPKPSIFRATRIRVWRITQVM-----DKIS---FQFEYLMAKDTFEWITLD------ 436 Query: 1017 VVISNNQTDQIILLSMSLKNMAEEYVCYKSSNSSLSDVENGEK-----MMNTSKFTSNDS 1181 TDQ IL+S+ L+++ E + Y+ +N ++ EK + S+ D Sbjct: 437 -------TDQSILMSLLLQSIGSE-ILYEHNNMTIEQQVMKEKHSKGNYVEKSEKLPRDP 488 Query: 1182 YKPAIMCSLYIGDGD 1226 KP I+ + D D Sbjct: 489 KKPIIVLKNEVEDTD 503
>sp|Q7NAK6|GIDA_MYCGA tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA (Glucose-inhibited division protein A) Length = 622 Score = 32.7 bits (73), Expect = 2.5 Identities = 31/114 (27%), Positives = 56/114 (49%), Gaps = 4/114 (3%) Frame = +3 Query: 393 SSKFSLFFMVVHPDMKFSIIRKLQDFEYPYVSLYEIQKLCNCRIVVRKSVWNIELESEID 572 ++ FSL+ + P++K + KL +F+Y S Y+++ L N I V+ + I+ ES I Sbjct: 503 NTNFSLYEFLKRPEIKLIELLKLIEFDY---SNYDLELLKNIEITVKYEGY-IKKESRIV 558 Query: 573 SHNSGLEILFAQVSAEIVNGWISIPDEARMNRIQNL----IERKARKETLQLAR 722 + LE I IP + +++QNL I++ + + L LA+ Sbjct: 559 NSLKNLES-------------IKIPKDLIYDKVQNLSIEAIDKLNKIKPLNLAQ 599
>sp|Q9Z6S3|TRMB_CHLPN tRNA (guanine-N(7)-)-methyltransferase (tRNA(m7G46)-methyltransferase) Length = 224 Score = 32.3 bits (72), Expect = 3.3 Identities = 13/36 (36%), Positives = 18/36 (50%) Frame = +1 Query: 295 WMARNTNLMSGALIIPKQYLRHLLMS*VYHEKFPQN 402 W R + G L +P+ Y H S YH++F QN Sbjct: 12 WKERRPCIQDGVLYVPRHYFEHQNFSTSYHQEFFQN 47
>sp|Q92J94|RS7_RICCN 30S ribosomal protein S7 Length = 160 Score = 31.6 bits (70), Expect = 5.6 Identities = 16/47 (34%), Positives = 25/47 (53%) Frame = +3 Query: 198 KFNSDELHDFLKTCQKETKKIVSEPVKLSVYLMDGQKHELDVWSTDN 338 K+NS L F+ KE KK ++E + S + +KH +D + T N Sbjct: 17 KYNSILLSRFINNIMKEGKKALAEKIVYSAFNKIEKKHRVDPYQTFN 63
>sp|P54939|TLN1_CHICK Talin-1 Length = 2541 Score = 31.2 bits (69), Expect = 7.3 Identities = 24/79 (30%), Positives = 38/79 (48%), Gaps = 4/79 (5%) Frame = +3 Query: 675 NLIERKARKETLQLARTLSYYG---YMQLHGTSVCNRPLPNTMAIVSTGNFEIVFR-DEH 842 N+ E +A+ ++LAR+L YG ++ N+ +P + I E V R DE Sbjct: 288 NMSEIEAKVRYVKLARSLKTYGVSFFLVKEKMKGKNKLVPRLLGITK----ECVMRVDEK 343 Query: 843 DDHVLMKYYLTRIKAWKIS 899 V+ ++ LT IK W S Sbjct: 344 TKEVIQEWSLTNIKRWAAS 362
>sp|P25386|USO1_YEAST Intracellular protein transport protein USO1 (Int-1) Length = 1790 Score = 31.2 bits (69), Expect = 7.3 Identities = 30/117 (25%), Positives = 51/117 (43%), Gaps = 12/117 (10%) Frame = +3 Query: 813 NFEIVFRDEHDDHVLMKYYLTRI-------KAWKISTAL-----KEPLNDDINQSVNQSQ 956 N + + E D+ L K Y + ++I TAL +EP+N + V + Q Sbjct: 677 NEDSILTPELDETGLPKVYFSTYFIQLFNENIYRIRTALSHDPDEEPINKISFEEVEKLQ 736 Query: 957 ALSPTLTFEYLDIETKKLDEVVISNNQTDQIILLSMSLKNMAEEYVCYKSSNSSLSD 1127 L E ++T+ N T+++I L+ K + E+Y SS+SSL + Sbjct: 737 RQCTKLKGEITSLQTETEST---HENLTEKLIALTNEHKELDEKYQILNSSHSSLKE 790
>sp|Q9H2U9|ADAM7_HUMAN ADAM 7 precursor (A disintegrin and metalloproteinase domain 7) (Sperm maturation-related glycoprotein GP-83) Length = 754 Score = 31.2 bits (69), Expect = 7.3 Identities = 21/81 (25%), Positives = 36/81 (44%) Frame = -3 Query: 672 VFYSFWLHRESISIHLQFLLKLEQREFQDQNYVNLFLILIRYSIHSFEQQFDNCKASGSH 493 + Y L+R+++ +HL L REF NY F + + Q D+C GS Sbjct: 59 LLYEIKLNRKTLVLHL-----LRSREFLGSNYSETFYSMKGGAFTRHPQIMDHCFYQGSI 113 Query: 492 KEKHRDTQNLATFE*LRTSYR 430 ++ +++T LR +R Sbjct: 114 VHEYDSAASISTCNGLRGFFR 134
>sp|Q28475|ADAM7_MACFA ADAM 7 precursor (A disintegrin and metalloproteinase domain 7) (Epididymal apical protein I) (EAP I) Length = 776 Score = 30.8 bits (68), Expect = 9.5 Identities = 21/81 (25%), Positives = 36/81 (44%) Frame = -3 Query: 672 VFYSFWLHRESISIHLQFLLKLEQREFQDQNYVNLFLILIRYSIHSFEQQFDNCKASGSH 493 + Y L+R+++ +HL L REF NY F + + Q D+C GS Sbjct: 59 LMYEIKLNRKTLVLHL-----LRSREFLGSNYSETFYSMKGEAFTRHLQIMDHCFYQGSI 113 Query: 492 KEKHRDTQNLATFE*LRTSYR 430 ++ +++T LR +R Sbjct: 114 VHEYDSAASISTCNGLRGFFR 134
>sp|Q92G13|HEMK_RICCN Bifunctional methyltransferase [Includes: HemK protein homolog (M.RcoHemKP); tRNA (guanine-N(7)-)-methyltransferase (tRNA(m7G46)-methyltransferase)] Length = 524 Score = 30.8 bits (68), Expect = 9.5 Identities = 26/107 (24%), Positives = 52/107 (48%), Gaps = 5/107 (4%) Frame = +3 Query: 582 SGLEILFAQVSAEIVNG-WISIPDEARMNRIQNLIERKARKETLQLARTLSYYGYMQLHG 758 + L+++ ++ + ++G +I PD N+ + +R KE L+L + + Sbjct: 404 NNLDLILNEIPSNSLDGIYILFPDPWIKNKQKK--KRIFNKERLKLLQDKLKDNGNLVFA 461 Query: 759 TSVCNRPLPNTMAIVSTGNFEIVFRDE----HDDHVLMKYYLTRIKA 887 + + N I GNFEI+ +++ HD++V+ KY+ IKA Sbjct: 462 SDIENYFYEAIELIQQNGNFEIINKNDYLKPHDNYVITKYHQKAIKA 508
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 152,735,483
Number of Sequences: 369166
Number of extensions: 3135940
Number of successful extensions: 7939
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7522
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7929
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 15985774220
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail