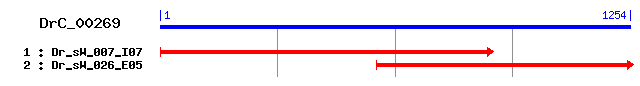
DrC_00269
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00269 (1254 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P48643|TCPE_HUMAN T-complex protein 1, epsilon subunit (... 416 e-116 sp|Q5RF02|TCPE_PONPY T-complex protein 1, epsilon subunit (... 414 e-115 sp|P80316|TCPE_MOUSE T-complex protein 1, epsilon subunit (... 413 e-115 sp|Q9UTM4|TCPE_SCHPO T-complex protein 1, epsilon subunit (... 382 e-105 sp|P47209|TCPE_CAEEL T-complex protein 1, epsilon subunit (... 378 e-104 sp|O04450|TCPE_ARATH T-complex protein 1, epsilon subunit (... 362 e-99 sp|P54411|TCPE2_AVESA T-complex protein 1, epsilon subunit ... 361 2e-99 sp|P40412|TCPE1_AVESA T-complex protein 1, epsilon subunit ... 357 3e-98 sp|P40413|TCPE_YEAST T-complex protein 1, epsilon subunit (... 337 4e-92 sp|Q58405|THS_METJA Thermosome subunit (Chaperonin subunit) 217 6e-56
>sp|P48643|TCPE_HUMAN T-complex protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon) Length = 541 Score = 416 bits (1068), Expect = e-116 Identities = 200/276 (72%), Positives = 241/276 (87%) Frame = +1 Query: 25 LKLI*SHIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKL 204 L+ + SHIMAAK V+N +RTSLGP GLDK+MV+ D D+TVTNDG TI+ MMDV+H+IAKL Sbjct: 31 LEALKSHIMAAKAVANTMRTSLGPNGLDKMMVDKDGDVTVTNDGATILSMMDVDHQIAKL 90 Query: 205 MVELSKSQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDK 384 MVELSKSQDDEIGDGTTGVVVLAG+LLE+AEQLLDRGIHPIRI+DGYE AA++ ++ LDK Sbjct: 91 MVELSKSQDDEIGDGTTGVVVLAGALLEEAEQLLDRGIHPIRIADGYEQAARVAIEHLDK 150 Query: 385 ISDTFVFSLENKEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIK 564 ISD+ + +++ EPLIQTA TTLGSK+VN CHRQ AE+AV A+L+VAD+ER+DVDFELIK Sbjct: 151 ISDSVLVDIKDTEPLIQTAKTTLGSKVVNSCHRQMAEIAVNAVLTVADMERRDVDFELIK 210 Query: 565 VDGKVGGTLSDSILVKGVIVDKDFSHPQMPKSIKDAKIAILTCAFEPPKPKTKHNLYVET 744 V+GKVGG L D+ L+KGVIVDKDFSHPQMPK ++DAKIAILTC FEPPKPKTKH L V + Sbjct: 211 VEGKVGGRLEDTKLIKGVIVDKDFSHPQMPKKVEDAKIAILTCPFEPPKPKTKHKLDVTS 270 Query: 745 VKDFENLAKYEHDKFVEMVDQVKASGANLVICQWGF 852 V+D++ L KYE +KF EM+ Q+K +GANL ICQWGF Sbjct: 271 VEDYKALQKYEKEKFEEMIQQIKETGANLAICQWGF 306
Score = 214 bits (544), Expect = 5e-55
Identities = 103/122 (84%), Positives = 114/122 (93%)
Frame = +2
Query: 848 GFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVRELSF 1027
GFDDEANHLL Q NLPAVRWVGGPEIELIAIATGGRIVPRF ELT EKLG AG+V+E+SF
Sbjct: 305 GFDDEANHLLLQNNLPAVRWVGGPEIELIAIATGGRIVPRFSELTAEKLGFAGLVQEISF 364
Query: 1028 GTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVYGGGA 1207
GTTKDKMLVIE+CKNS+AVT FIRGG++MI++EAKRSLHDALCV+RNL+RD RVVYGGGA
Sbjct: 365 GTTKDKMLVIEQCKNSRAVTIFIRGGNKMIIEEAKRSLHDALCVIRNLIRDNRVVYGGGA 424
Query: 1208 PE 1213
E
Sbjct: 425 AE 426
>sp|Q5RF02|TCPE_PONPY T-complex protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon) Length = 541 Score = 414 bits (1064), Expect = e-115 Identities = 199/276 (72%), Positives = 241/276 (87%) Frame = +1 Query: 25 LKLI*SHIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKL 204 L+ + SHIMAAK V+N +RTSLGP GLDK+MV+ D D+TVTNDG TI+ MMDV+H+IAKL Sbjct: 31 LEALKSHIMAAKAVANTMRTSLGPNGLDKMMVDKDGDVTVTNDGATILSMMDVDHQIAKL 90 Query: 205 MVELSKSQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDK 384 MVELSKSQDDEIGDGTTGVVVLAG+LLE+AEQLLDRGIHPIRI+DGYE AA++ ++ LDK Sbjct: 91 MVELSKSQDDEIGDGTTGVVVLAGALLEEAEQLLDRGIHPIRIADGYEQAARVAIEHLDK 150 Query: 385 ISDTFVFSLENKEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIK 564 ISD+ + +++ EPLIQTA TTLGSK+VN CHRQ AE+AV A+L+VAD+ER+DVDFELIK Sbjct: 151 ISDSVLVDIKDTEPLIQTAKTTLGSKVVNSCHRQMAEIAVNAVLTVADMERRDVDFELIK 210 Query: 565 VDGKVGGTLSDSILVKGVIVDKDFSHPQMPKSIKDAKIAILTCAFEPPKPKTKHNLYVET 744 V+GKVGG L D+ L+KGVIVDKDFSHP+MPK ++DAKIAILTC FEPPKPKTKH L V + Sbjct: 211 VEGKVGGRLEDTKLIKGVIVDKDFSHPRMPKKVEDAKIAILTCPFEPPKPKTKHKLDVTS 270 Query: 745 VKDFENLAKYEHDKFVEMVDQVKASGANLVICQWGF 852 V+D++ L KYE +KF EM+ Q+K +GANL ICQWGF Sbjct: 271 VEDYKALQKYEKEKFEEMIQQIKETGANLAICQWGF 306
Score = 214 bits (544), Expect = 5e-55
Identities = 103/122 (84%), Positives = 114/122 (93%)
Frame = +2
Query: 848 GFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVRELSF 1027
GFDDEANHLL Q NLPAVRWVGGPEIELIAIATGGRIVPRF ELT EKLG AG+V+E+SF
Sbjct: 305 GFDDEANHLLLQNNLPAVRWVGGPEIELIAIATGGRIVPRFSELTAEKLGFAGLVQEISF 364
Query: 1028 GTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVYGGGA 1207
GTTKDKMLVIE+CKNS+AVT FIRGG++MI++EAKRSLHDALCV+RNL+RD RVVYGGGA
Sbjct: 365 GTTKDKMLVIEQCKNSRAVTIFIRGGNKMIIEEAKRSLHDALCVIRNLIRDNRVVYGGGA 424
Query: 1208 PE 1213
E
Sbjct: 425 AE 426
>sp|P80316|TCPE_MOUSE T-complex protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon) Length = 541 Score = 413 bits (1062), Expect = e-115 Identities = 199/276 (72%), Positives = 237/276 (85%) Frame = +1 Query: 25 LKLI*SHIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKL 204 L+ + SHIMAAK V+N +RTSLGP GLDK+MV+ D D+T+TNDG TI+ MMDV+H+IAKL Sbjct: 31 LEALKSHIMAAKAVANTMRTSLGPNGLDKMMVDKDGDVTITNDGATILSMMDVDHQIAKL 90 Query: 205 MVELSKSQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDK 384 MVELSKSQDDEIGDGTTGVVVLAG+LLE+AEQLLDRGIHPIRI+DGYE AA++ + LDK Sbjct: 91 MVELSKSQDDEIGDGTTGVVVLAGALLEEAEQLLDRGIHPIRIADGYEQAARIAIQHLDK 150 Query: 385 ISDTFVFSLENKEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIK 564 ISD + + N EPLIQTA TTLGSK++N CHRQ AE+AV A+L+VAD+ER+DVDFELIK Sbjct: 151 ISDKVLVDINNPEPLIQTAKTTLGSKVINSCHRQMAEIAVNAVLTVADMERRDVDFELIK 210 Query: 565 VDGKVGGTLSDSILVKGVIVDKDFSHPQMPKSIKDAKIAILTCAFEPPKPKTKHNLYVET 744 V+GKVGG L D+ L+KGVIVDKDFSHPQMPK + DAKIAILTC FEPPKPKTKH L V + Sbjct: 211 VEGKVGGRLEDTKLIKGVIVDKDFSHPQMPKKVVDAKIAILTCPFEPPKPKTKHKLDVMS 270 Query: 745 VKDFENLAKYEHDKFVEMVDQVKASGANLVICQWGF 852 V+D++ L KYE +KF EM+ Q+K +GANL ICQWGF Sbjct: 271 VEDYKALQKYEKEKFEEMIKQIKETGANLAICQWGF 306
Score = 212 bits (540), Expect = 2e-54
Identities = 103/122 (84%), Positives = 113/122 (92%)
Frame = +2
Query: 848 GFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVRELSF 1027
GFDDEANHLL Q LPAVRWVGGPEIELIAIATGGRIVPRF ELT EKLG AGVV+E+SF
Sbjct: 305 GFDDEANHLLLQNGLPAVRWVGGPEIELIAIATGGRIVPRFSELTSEKLGFAGVVQEISF 364
Query: 1028 GTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVYGGGA 1207
GTTKDKMLVIE+CKNS+AVT FIRGG++MI++EAKRSLHDALCV+RNL+RD RVVYGGGA
Sbjct: 365 GTTKDKMLVIEKCKNSRAVTIFIRGGNKMIIEEAKRSLHDALCVIRNLIRDNRVVYGGGA 424
Query: 1208 PE 1213
E
Sbjct: 425 AE 426
>sp|Q9UTM4|TCPE_SCHPO T-complex protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon) Length = 546 Score = 382 bits (981), Expect = e-105 Identities = 186/271 (68%), Positives = 228/271 (84%) Frame = +1 Query: 40 SHIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKLMVELS 219 SHI+A KTV+NI+RTSLGP+GLDKI+++PD +ITVTNDG TI++ M+VEH+IAKL+V+LS Sbjct: 38 SHILATKTVANIVRTSLGPRGLDKILISPDGEITVTNDGATILDQMEVEHQIAKLLVQLS 97 Query: 220 KSQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDKISDTF 399 KSQDDEIGDGTTGVVVLAG+LLEQAE L+D+GIHPIRI+DGYE A ++ + LD ISD Sbjct: 98 KSQDDEIGDGTTGVVVLAGALLEQAEALIDKGIHPIRIADGYEKACQVAVKHLDAISDVV 157 Query: 400 VFSLENKEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIKVDGKV 579 FS EN L ++A T+LGSK+V+K H FA +AV A+LSVAD++RKDVDFELIKVDGKV Sbjct: 158 DFSPENTTNLFRSAKTSLGSKVVSKAHDHFANIAVDAVLSVADLQRKDVDFELIKVDGKV 217 Query: 580 GGTLSDSILVKGVIVDKDFSHPQMPKSIKDAKIAILTCAFEPPKPKTKHNLYVETVKDFE 759 GG++ D+ LVKGV+VDKD SHPQMP I++AKIAILTC FEPPKPKTKH L + +V +FE Sbjct: 218 GGSVDDTKLVKGVVVDKDMSHPQMPHRIENAKIAILTCPFEPPKPKTKHKLDITSVSEFE 277 Query: 760 NLAKYEHDKFVEMVDQVKASGANLVICQWGF 852 L YE +KF EM+ VK +GANLVICQWGF Sbjct: 278 ALQAYEKEKFQEMIKHVKDAGANLVICQWGF 308
Score = 206 bits (524), Expect = 1e-52
Identities = 98/122 (80%), Positives = 113/122 (92%)
Frame = +2
Query: 848 GFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVRELSF 1027
GFDDEANHLL Q NLPAVRWVGGPEIELIAIAT GRIVPRFE+L+ +KLG+AG+VRE+SF
Sbjct: 307 GFDDEANHLLLQNNLPAVRWVGGPEIELIAIATNGRIVPRFEDLSSDKLGSAGIVREVSF 366
Query: 1028 GTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVYGGGA 1207
GTT+DK+LVIE+C NS+AVT F+RG ++MIVDEAKR+LHDALCVVRNL+RD RVVYGGGA
Sbjct: 367 GTTRDKILVIEKCANSRAVTVFVRGSNKMIVDEAKRALHDALCVVRNLIRDNRVVYGGGA 426
Query: 1208 PE 1213
E
Sbjct: 427 AE 428
>sp|P47209|TCPE_CAEEL T-complex protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon) Length = 542 Score = 378 bits (970), Expect = e-104 Identities = 182/271 (67%), Positives = 228/271 (84%) Frame = +1 Query: 40 SHIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKLMVELS 219 SHI+AA+ V+N LRTSLGP+GLDK++V+PD D+T+TNDG TIME MDV+H +AKLMVELS Sbjct: 37 SHILAARAVANTLRTSLGPRGLDKMLVSPDGDVTITNDGATIMEKMDVQHHVAKLMVELS 96 Query: 220 KSQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDKISDTF 399 KSQD EIGDGTTGVVVLAG+LLE+AE+L+DRGIHPI+I+DG+++A K L+ LD ISD F Sbjct: 97 KSQDHEIGDGTTGVVVLAGALLEEAEKLIDRGIHPIKIADGFDLACKKALETLDSISDKF 156 Query: 400 VFSLENKEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIKVDGKV 579 +EN+E L++TA T+LGSKIVN+ RQFAE+AV A+LSVADIE KDV+FE+IK++GKV Sbjct: 157 --PVENRERLVETAQTSLGSKIVNRSLRQFAEIAVDAVLSVADIESKDVNFEMIKMEGKV 214 Query: 580 GGTLSDSILVKGVIVDKDFSHPQMPKSIKDAKIAILTCAFEPPKPKTKHNLYVETVKDFE 759 GG L D+ILVKG+++DK SHPQMPK +K+AK+AILTC FEPPKPKTKH L + + +DF+ Sbjct: 215 GGRLEDTILVKGIVIDKTMSHPQMPKELKNAKVAILTCPFEPPKPKTKHKLDITSTEDFK 274 Query: 760 NLAKYEHDKFVEMVDQVKASGANLVICQWGF 852 L YE + F M+ QVK SGA L ICQWGF Sbjct: 275 ALRDYERETFETMIRQVKESGATLAICQWGF 305
Score = 195 bits (495), Expect = 3e-49
Identities = 90/122 (73%), Positives = 108/122 (88%)
Frame = +2
Query: 848 GFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVRELSF 1027
GFDDEANHLL +LPAVRWVGGPEIEL+AIAT RIVPRF EL++EKLG AG+VRE++F
Sbjct: 304 GFDDEANHLLQANDLPAVRWVGGPEIELLAIATNARIVPRFSELSKEKLGTAGLVREITF 363
Query: 1028 GTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVYGGGA 1207
G KD+ML IE+C N+KAVT F+RGG++MI+DEAKR+LHDALCV+RNLVRD R+VYGGG+
Sbjct: 364 GAAKDRMLSIEQCPNNKAVTIFVRGGNKMIIDEAKRALHDALCVIRNLVRDSRIVYGGGS 423
Query: 1208 PE 1213
E
Sbjct: 424 AE 425
>sp|O04450|TCPE_ARATH T-complex protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon) Length = 535 Score = 362 bits (930), Expect = e-99 Identities = 174/271 (64%), Positives = 222/271 (81%) Frame = +1 Query: 40 SHIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKLMVELS 219 ++I A K V+ ILR+SLGPKG+DK++ PD DIT+TNDG TI+E MDV+++IAKLMVELS Sbjct: 32 ANIAAGKAVARILRSSLGPKGMDKMLQGPDGDITITNDGATILEQMDVDNQIAKLMVELS 91 Query: 220 KSQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDKISDTF 399 +SQD EIGDGTTGVVV+AG+LLEQAE+ LDRGIHPIRI++GYEMA+++ ++ L++I+ F Sbjct: 92 RSQDYEIGDGTTGVVVMAGALLEQAERQLDRGIHPIRIAEGYEMASRVAVEHLERIAQKF 151 Query: 400 VFSLENKEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIKVDGKV 579 F + N EPL+QT MTTL SKIVN+C R AE+AV A+L+VAD+ER+DV+ +LIKV+GKV Sbjct: 152 EFDVNNYEPLVQTCMTTLSSKIVNRCKRSLAEIAVKAVLAVADLERRDVNLDLIKVEGKV 211 Query: 580 GGTLSDSILVKGVIVDKDFSHPQMPKSIKDAKIAILTCAFEPPKPKTKHNLYVETVKDFE 759 GG L D+ L+ G+++DKD SHPQMPK I+DA IAILTC FEPPKPKTKH + ++TV+ FE Sbjct: 212 GGKLEDTELIYGILIDKDMSHPQMPKQIEDAHIAILTCPFEPPKPKTKHKVDIDTVEKFE 271 Query: 760 NLAKYEHDKFVEMVDQVKASGANLVICQWGF 852 L K E F EMV + K GA LVICQWGF Sbjct: 272 TLRKQEQQYFDEMVQKCKDVGATLVICQWGF 302
Score = 196 bits (497), Expect = 2e-49
Identities = 93/122 (76%), Positives = 107/122 (87%)
Frame = +2
Query: 848 GFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVRELSF 1027
GFDDEANHLL +NLPAVRWVGG E+ELIAIATGGRIVPRF+ELT EKLG AGVVRE SF
Sbjct: 301 GFDDEANHLLMHRNLPAVRWVGGVELELIAIATGGRIVPRFQELTPEKLGKAGVVREKSF 360
Query: 1028 GTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVYGGGA 1207
GTTK++ML IE C NSKAVT FIRGG++M+++E KRS+HDALCV RNL+R+ +VYGGGA
Sbjct: 361 GTTKERMLYIEHCANSKAVTVFIRGGNKMMIEETKRSIHDALCVARNLIRNKSIVYGGGA 420
Query: 1208 PE 1213
E
Sbjct: 421 AE 422
>sp|P54411|TCPE2_AVESA T-complex protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon) (TCP-K36) Length = 535 Score = 361 bits (927), Expect = 2e-99 Identities = 175/271 (64%), Positives = 225/271 (83%) Frame = +1 Query: 40 SHIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKLMVELS 219 ++I A K+V+ ILRTSLGPKG+DK++ +PD D+T+TNDG TI+E+MDV+++IAKLMVELS Sbjct: 32 ANIAAGKSVARILRTSLGPKGMDKMLQSPDGDVTITNDGATILELMDVDNQIAKLMVELS 91 Query: 220 KSQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDKISDTF 399 +SQD +IGDGTTGVVV+AGSLLEQAE+LL+RGIHPIR+++GYEMA+++ +D L+ IS + Sbjct: 92 RSQDYDIGDGTTGVVVMAGSLLEQAEKLLERGIHPIRVAEGYEMASRIAVDHLESISTKY 151 Query: 400 VFSLENKEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIKVDGKV 579 FS + EPL+QT MTTL SKIV++C R AE+AV A+L+VAD+ERKDV+ +LIKV+GKV Sbjct: 152 EFSATDIEPLVQTCMTTLSSKIVSRCKRALAEIAVKAVLAVADLERKDVNLDLIKVEGKV 211 Query: 580 GGTLSDSILVKGVIVDKDFSHPQMPKSIKDAKIAILTCAFEPPKPKTKHNLYVETVKDFE 759 GG L D+ LV+G+IVDKD SHPQMPK I+DA IAILTC FEPPKPKTKH + ++TV+ F+ Sbjct: 212 GGKLEDTELVQGIIVDKDMSHPQMPKRIEDAHIAILTCPFEPPKPKTKHKVDIDTVEKFQ 271 Query: 760 NLAKYEHDKFVEMVDQVKASGANLVICQWGF 852 L E F EMV + K GA LVICQWGF Sbjct: 272 TLRGQEQKYFDEMVQKCKDVGATLVICQWGF 302
Score = 190 bits (483), Expect = 6e-48
Identities = 90/122 (73%), Positives = 107/122 (87%)
Frame = +2
Query: 848 GFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVRELSF 1027
GFDDEANHLL Q+ LPAVRWVGG E+ELIAIATGGRIVPRF+EL+ EKLG AG+VRE SF
Sbjct: 301 GFDDEANHLLMQRELPAVRWVGGVELELIAIATGGRIVPRFQELSTEKLGKAGLVREKSF 360
Query: 1028 GTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVYGGGA 1207
GTTKD+ML IE+C NSKAVT FIRGG++M+++E KRS+HDALCV RNL+ + +VYGGG+
Sbjct: 361 GTTKDRMLYIEKCANSKAVTIFIRGGNKMMIEETKRSIHDALCVARNLIINNSIVYGGGS 420
Query: 1208 PE 1213
E
Sbjct: 421 AE 422
>sp|P40412|TCPE1_AVESA T-complex protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon) (TCP-K19) Length = 535 Score = 357 bits (917), Expect = 3e-98 Identities = 172/271 (63%), Positives = 224/271 (82%) Frame = +1 Query: 40 SHIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKLMVELS 219 ++I AAK ++ ILRTSLGPKG+DK++ +PD D+T+TNDG TI+E+MDV+++IAKL+VELS Sbjct: 32 ANIAAAKAIARILRTSLGPKGMDKMLQSPDGDVTITNDGATILELMDVDNQIAKLLVELS 91 Query: 220 KSQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDKISDTF 399 +SQD +IGDGTTGVVV+AG+LLEQAE+LL+RGIHPIR+++GYEMA+++ +D L+ IS + Sbjct: 92 RSQDYDIGDGTTGVVVMAGALLEQAEKLLERGIHPIRVAEGYEMASRIAVDHLESISTKY 151 Query: 400 VFSLENKEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIKVDGKV 579 FS + EPL+QT MTTL SKIV++C R AE++V A+L+VAD+ERKDV+ +LIKV+GKV Sbjct: 152 EFSATDIEPLVQTCMTTLSSKIVSRCKRALAEISVKAVLAVADLERKDVNLDLIKVEGKV 211 Query: 580 GGTLSDSILVKGVIVDKDFSHPQMPKSIKDAKIAILTCAFEPPKPKTKHNLYVETVKDFE 759 GG L D+ LV+G+IVDKD SHPQMPK I DA IAILTC FEPPKPKTKH + ++TV+ F+ Sbjct: 212 GGKLEDTELVEGIIVDKDMSHPQMPKRIYDAHIAILTCPFEPPKPKTKHKVDIDTVEKFQ 271 Query: 760 NLAKYEHDKFVEMVDQVKASGANLVICQWGF 852 L E F EMV + K GA LVICQWGF Sbjct: 272 TLRGQEQKYFDEMVQKCKDVGATLVICQWGF 302
Score = 190 bits (483), Expect = 6e-48
Identities = 90/122 (73%), Positives = 107/122 (87%)
Frame = +2
Query: 848 GFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVRELSF 1027
GFDDEANHLL Q+ LPAVRWVGG E+ELIAIATGGRIVPRF+EL+ EKLG AG+VRE SF
Sbjct: 301 GFDDEANHLLMQRELPAVRWVGGVELELIAIATGGRIVPRFQELSTEKLGKAGLVREKSF 360
Query: 1028 GTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVYGGGA 1207
GTTKD+ML IE+C NSKAVT FIRGG++M+++E KRS+HDALCV RNL+ + +VYGGG+
Sbjct: 361 GTTKDRMLYIEKCANSKAVTIFIRGGNKMMIEETKRSIHDALCVARNLIINNSIVYGGGS 420
Query: 1208 PE 1213
E
Sbjct: 421 AE 422
>sp|P40413|TCPE_YEAST T-complex protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon) Length = 551 Score = 337 bits (864), Expect = 4e-92 Identities = 162/279 (58%), Positives = 227/279 (81%), Gaps = 8/279 (2%) Frame = +1 Query: 40 SHIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKLMVELS 219 SHI+AA++V++I++TSLGP+GLDKI+++PD +IT+TNDG TI+ M++++EIAKL+V+LS Sbjct: 38 SHILAARSVASIIKTSLGPRGLDKILISPDGEITITNDGATILSQMELDNEIAKLLVQLS 97 Query: 220 KSQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDKISDTF 399 KSQDDEIGDGTTGVVVLA +LL+QA +L+ +GIHPI+I++G++ AAKL + +L++ D Sbjct: 98 KSQDDEIGDGTTGVVVLASALLDQALELIQKGIHPIKIANGFDEAAKLAISKLEETCDDI 157 Query: 400 VFSLEN--KEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIKVDG 573 S + ++ L++ A T+LGSKIV+K H +FAEMAV A+++V D +RKDVDF+LIK+ G Sbjct: 158 SASNDELFRDFLLRAAKTSLGSKIVSKDHDRFAEMAVEAVINVMDKDRKDVDFDLIKMQG 217 Query: 574 KVGGTLSDSILVKGVIVDKDFSHPQMPKSI------KDAKIAILTCAFEPPKPKTKHNLY 735 +VGG++SDS L+ GVI+DKDFSHPQMPK + K+AILTC FEPPKPKTKH L Sbjct: 218 RVGGSISDSKLINGVILDKDFSHPQMPKCVLPKEGSDGVKLAILTCPFEPPKPKTKHKLD 277 Query: 736 VETVKDFENLAKYEHDKFVEMVDQVKASGANLVICQWGF 852 + +V++++ L YE DKF EM+D VK +GA++VICQWGF Sbjct: 278 ISSVEEYQKLQTYEQDKFKEMIDDVKKAGADVVICQWGF 316
Score = 181 bits (459), Expect = 4e-45
Identities = 84/122 (68%), Positives = 104/122 (85%)
Frame = +2
Query: 848 GFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVRELSF 1027
GFDDEANHLL Q +LPAVRWVGG E+E IAI+T GRIVPRF++L+++KLG + E F
Sbjct: 315 GFDDEANHLLLQNDLPAVRWVGGQELEHIAISTNGRIVPRFQDLSKDKLGTCSRIYEQEF 374
Query: 1028 GTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVYGGGA 1207
GTTKD+ML+IE+ K +K VTCF+RG ++MIVDEA+R+LHD+LCVVRNLV+D RVVYGGGA
Sbjct: 375 GTTKDRMLIIEQSKETKTVTCFVRGSNKMIVDEAERALHDSLCVVRNLVKDSRVVYGGGA 434
Query: 1208 PE 1213
E
Sbjct: 435 AE 436
>sp|Q58405|THS_METJA Thermosome subunit (Chaperonin subunit) Length = 542 Score = 217 bits (552), Expect = 6e-56 Identities = 112/269 (41%), Positives = 174/269 (64%) Frame = +1 Query: 43 HIMAAKTVSNILRTSLGPKGLDKIMVNPDNDITVTNDGRTIMEMMDVEHEIAKLMVELSK 222 +I+A + ++ +RT+LGPKG+DK++V+ DI VTNDG TI++ M VEH AK+++E++K Sbjct: 27 NILAGRIIAETVRTTLGPKGMDKMLVDELGDIVVTNDGVTILKEMSVEHPAAKMLIEVAK 86 Query: 223 SQDDEIGDGTTGVVVLAGSLLEQAEQLLDRGIHPIRISDGYEMAAKLCLDELDKISDTFV 402 +Q+ E+GDGTT VV+AG LL +AE+LLD+ IHP I +GYEMA ++EL I+ Sbjct: 87 TQEKEVGDGTTTAVVIAGELLRKAEELLDQNIHPSVIINGYEMARNKAVEELKSIAKE-- 144 Query: 403 FSLENKEPLIQTAMTTLGSKIVNKCHRQFAEMAVGAILSVADIERKDVDFELIKVDGKVG 582 E+ E L + AMT++ K K Q AE+ V A+ +V D E VD +LIKV+ K G Sbjct: 145 VKPEDTEMLKKIAMTSITGKGAEKAREQLAEIVVEAVRAVVDEETGKVDKDLIKVEKKEG 204 Query: 583 GTLSDSILVKGVIVDKDFSHPQMPKSIKDAKIAILTCAFEPPKPKTKHNLYVETVKDFEN 762 + ++ L++GV++DK+ +PQMPK +++AKIA+L C E + +T + + Sbjct: 205 APIEETKLIRGVVIDKERVNPQMPKKVENAKIALLNCPIEVKETETDAEIRITDPAKLME 264 Query: 763 LAKYEHDKFVEMVDQVKASGANLVICQWG 849 + E +MV+++ A+GAN+V CQ G Sbjct: 265 FIEQEEKMIKDMVEKIAATGANVVFCQKG 293
Score = 97.4 bits (241), Expect = 7e-20
Identities = 53/126 (42%), Positives = 77/126 (61%)
Frame = +2
Query: 836 FVSGGFDDEANHLLYQKNLPAVRWVGGPEIELIAIATGGRIVPRFEELTREKLGNAGVVR 1015
F G DD A H L +K + AVR V ++E +A ATG RIV + ++LT E LG AG+V
Sbjct: 289 FCQKGIDDLAQHYLAKKGILAVRRVKKSDMEKLAKATGARIVTKIDDLTPEDLGEAGLVE 348
Query: 1016 ELSFGTTKDKMLVIEECKNSKAVTCFIRGGSRMIVDEAKRSLHDALCVVRNLVRDPRVVY 1195
E D M+ +E+CK+ KAVT RG + +V+E R++ DA+ VV+ + + ++V
Sbjct: 349 ERK--VAGDAMIFVEQCKHPKAVTILARGSTEHVVEEVARAIDDAIGVVKCALEEGKIVA 406
Query: 1196 GGGAPE 1213
GGGA E
Sbjct: 407 GGGATE 412
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 131,406,840
Number of Sequences: 369166
Number of extensions: 2434025
Number of successful extensions: 8079
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7122
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7578
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14433897200
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail