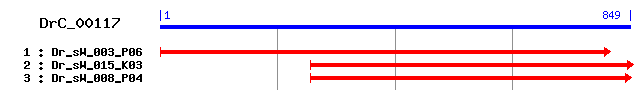
DrC_00117
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00117 (849 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9BY32|ITPA_HUMAN Inosine triphosphate pyrophosphatase (... 127 5e-29 sp|Q9D892|ITPA_MOUSE Inosine triphosphate pyrophosphatase (... 123 6e-28 sp|P47119|HAM1_YEAST HAM1 protein 101 2e-21 sp|Q57679|NTPA_METJA Nucleoside-triphosphatase (Nucleoside ... 85 3e-16 sp|Q8TV07|HAM1_METKA HAM1 protein homolog 79 1e-14 sp|Q5JEX8|HAM1_PYRKO HAM1 protein homolog 73 1e-12 sp|Q8TJS1|HAM1_METAC HAM1 protein homolog 71 4e-12 sp|Q8A327|HAM1_BACTN HAM1 protein homolog 70 1e-11 sp|Q8U446|HAM1_PYRFU HAM1 protein homolog 69 1e-11 sp|Q8PZ91|HAM1_METMA HAM1 protein homolog 68 3e-11
>sp|Q9BY32|ITPA_HUMAN Inosine triphosphate pyrophosphatase (ITPase) (Inosine triphosphatase) (Putative oncogene protein hlc14-06-p) Length = 194 Score = 127 bits (318), Expect = 5e-29 Identities = 73/164 (44%), Positives = 96/164 (58%) Frame = +2 Query: 275 IMSQSIDIPEYQEEIIEISLLKAKYAYSIIKEPVFVEDTSLCFNALYGMPGPYIKWFVHG 454 +++Q ID+PEYQ E EIS+ K + A ++ PV VEDT LCFNAL G+PGPYIKWF+ Sbjct: 35 LVAQKIDLPEYQGEPDEISIQKCQEAVRQVQGPVLVEDTCLCFNALGGLPGPYIKWFL-- 92 Query: 455 NLDEKFKKLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFIGKMLGEIVEPRGPMD 634 EK K +GL ++L GF D A A+ + Q + F G+ G IV PRG D Sbjct: 93 ---EKLKP-EGLHQLLAGFEDKSAYALCTFALSTGDPSQPVRLFRGRTSGRIVAPRGCQD 148 Query: 635 FDWDCVFQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEFKKF 766 F WD FQP QTYAEM KN +S+R+ A E +++ Sbjct: 149 FGWDPCFQP----DGYEQTYAEMPKAEKNAVSHRFRALLELQEY 188
>sp|Q9D892|ITPA_MOUSE Inosine triphosphate pyrophosphatase (ITPase) (Inosine triphosphatase) Length = 198 Score = 123 bits (309), Expect = 6e-28 Identities = 72/162 (44%), Positives = 94/162 (58%) Frame = +2 Query: 281 SQSIDIPEYQEEIIEISLLKAKYAYSIIKEPVFVEDTSLCFNALYGMPGPYIKWFVHGNL 460 +Q ID+PEYQ E EIS+ K + A ++ PV VEDT LCFNAL G+PGPYIKWF+ Sbjct: 37 AQKIDLPEYQGEPDEISIQKCREAARQVQGPVLVEDTCLCFNALGGLPGPYIKWFL---- 92 Query: 461 DEKFKKLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFIGKMLGEIVEPRGPMDFD 640 +K K +GL ++L GF D A A+ + Q F G+ G+IV PRG DF Sbjct: 93 -QKLKP-EGLHQLLAGFEDKSAYALCTFALSTGDPSQPVLLFRGQTSGQIVMPRGSRDFG 150 Query: 641 WDCVFQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEFKKF 766 WD FQP QTYAEM KN IS+R+ A + +++ Sbjct: 151 WDPCFQP----DGYEQTYAEMPKSEKNTISHRFRALHKLQEY 188
>sp|P47119|HAM1_YEAST HAM1 protein Length = 197 Score = 101 bits (252), Expect = 2e-21 Identities = 66/171 (38%), Positives = 98/171 (57%), Gaps = 3/171 (1%) Frame = +2 Query: 275 IMSQSIDIPEYQE-EIIEISLLKAKYAYSIIKE--PVFVEDTSLCFNALYGMPGPYIKWF 445 ++++++D+ E Q+ ++ I+L K K A + + + PVFVEDT+L F+ G+PG YIKWF Sbjct: 36 LINEALDLEELQDTDLNAIALAKGKQAVAALGKGKPVFVEDTALRFDEFNGLPGAYIKWF 95 Query: 446 VHGNLDEKFKKLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFIGKMLGEIVEPRG 625 + K L+ + KML F + AEAVT I + D + + F G G+IV RG Sbjct: 96 L------KSMGLEKIVKMLEPFENKNAEAVTTICFADSR--GEYHFFQGITRGKIVPSRG 147 Query: 626 PMDFDWDCVFQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEFKKFLLEN 778 P F WD +F+P TYAEM KN IS+R A +FK++L +N Sbjct: 148 PTTFGWDSIFEPF---DSHGLTYAEMSKDAKNAISHRGKAFAQFKEYLYQN 195
>sp|Q57679|NTPA_METJA Nucleoside-triphosphatase (Nucleoside triphosphate phosphohydrolase) (NTPase) Length = 193 Score = 84.7 bits (208), Expect = 3e-16 Identities = 56/162 (34%), Positives = 89/162 (54%) Frame = +2 Query: 290 IDIPEYQEEIIEISLLKAKYAYSIIKEPVFVEDTSLCFNALYGMPGPYIKWFVHGNLDEK 469 I PE Q + E++ AK+ Y+I+K+PV VED+ AL G PG Y K FV + Sbjct: 41 ISYPEIQGTLEEVAEFGAKWVYNILKKPVIVEDSGFFVEALNGFPGTYSK-FVQETIGN- 98 Query: 470 FKKLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFIGKMLGEIVEPRGPMDFDWDC 649 +G+ K+L G ++ A T+I Y D ++ K + + E + +G F +D Sbjct: 99 ----EGILKLLEGKDNRNAYFKTVIGYCDENGVRLFKGIVKGRVSEEIRSKG-YGFAYDS 153 Query: 650 VFQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEFKKFLLE 775 +F P ++ +T+AEM + K++IS+R A +EFKKFLL+ Sbjct: 154 IFIP----EEEERTFAEMTTEEKSQISHRKKAFEEFKKFLLD 191
>sp|Q8TV07|HAM1_METKA HAM1 protein homolog Length = 188 Score = 79.3 bits (194), Expect = 1e-14 Identities = 55/167 (32%), Positives = 86/167 (51%), Gaps = 4/167 (2%) Frame = +2 Query: 290 IDIPEYQEEIIE-ISLLKAKYAYSIIKEPVFVEDTSLCFNALYGMPGPYIKWFVH--GNL 460 +D PE Q + +E I+ A+Y + +PV VED+ L AL G PGPY + GN Sbjct: 32 LDYPELQSDSLEEIAAYGARYCAESLGQPVIVEDSGLFIEALNGFPGPYSAYVFDTIGN- 90 Query: 461 DEKFKKLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFIGKMLGEIV-EPRGPMDF 637 +G+ K+L G + +AE ++++ Y +P P F G++ G I EPRG F Sbjct: 91 -------EGILKLLEGEENRKAEFISVVGYCEPG--GRPVTFTGEIRGRIAEEPRGEEGF 141 Query: 638 DWDCVFQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEFKKFLLEN 778 +D +F P + T+AE+ + K KIS+R A + F ++ N Sbjct: 142 GYDPIFIPEGED----STFAELGVEEKCKISHRTKALERFAEWYKNN 184
>sp|Q5JEX8|HAM1_PYRKO HAM1 protein homolog Length = 184 Score = 72.8 bits (177), Expect = 1e-12 Identities = 53/165 (32%), Positives = 87/165 (52%), Gaps = 2/165 (1%) Frame = +2 Query: 290 IDIPEYQEEIIE-ISLLKAKYAYSIIKEPVFVEDTSLCFNALYGMPGPYIKWFVHGNLDE 466 + PE Q + +E ++ AK+ + P F++D+ L AL G PG Y + Sbjct: 32 VSYPEIQADTLEEVAEYGAKWLAQRVDGPFFLDDSGLFVEALKGFPGVYSAYVY------ 85 Query: 467 KFKKLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFIGKMLGEIV-EPRGPMDFDW 643 K QG+ K+L G + +A ++I+Y+D ++ F G++ G+I EPRG F + Sbjct: 86 KTIGYQGILKLLQGEKNRKAHFKSVIAYWDGEL----HIFTGRVDGKIATEPRGSGGFGF 141 Query: 644 DCVFQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEFKKFLLEN 778 D +F P + +T+AEM + KN+IS+R A +EF +L EN Sbjct: 142 DPIFIP----EGFDRTFAEMTTEEKNRISHRGRALREFANWLKEN 182
>sp|Q8TJS1|HAM1_METAC HAM1 protein homolog Length = 184 Score = 70.9 bits (172), Expect = 4e-12 Identities = 53/163 (32%), Positives = 85/163 (52%), Gaps = 2/163 (1%) Frame = +2 Query: 275 IMSQSIDIPEYQEEIIE-ISLLKAKYAYSIIKEPVFVEDTSLCFNALYGMPGPYIKWFVH 451 ++ + PE QE+ +E I+ A+Y + + PV V+D+ + NAL G PGPY ++ Sbjct: 28 VIQEKNGYPELQEDELEPIAAHGAQYVANKLNMPVMVDDSGIFINALNGFPGPYSRF--- 84 Query: 452 GNLDEKFKKLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFIGKMLGEIV-EPRGP 628 +++K L+ L KM+ G D A T+I Y +P ++P F G + G+I E RG Sbjct: 85 --VEDKLGNLKVL-KMMEGEEDRTAYFKTVIGYCEPG--KEPLVFPGVVEGKIAYEERGT 139 Query: 629 MDFDWDCVFQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEF 757 F +D +F+ T+ E+ KNK+S+R A EF Sbjct: 140 GGFGYDPIFEYQG------LTFGELGDTEKNKVSHRRRAVDEF 176
>sp|Q8A327|HAM1_BACTN HAM1 protein homolog Length = 193 Score = 69.7 bits (169), Expect = 1e-11 Identities = 54/162 (33%), Positives = 83/162 (51%), Gaps = 3/162 (1%) Frame = +2 Query: 293 DIPEYQEEIIEISLLKAKYAYSIIKEPVFVEDTSLCFNALYGMPGPYIKWFVHGNLDEKF 472 DIPE E + +LLK+ + Y + F +DT L AL G PG Y + G + Sbjct: 39 DIPETAETLEGNALLKSSFIYRNYQLDCFADDTGLEVEALNGAPGVYSARYAEGEGHDAQ 98 Query: 473 KKLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFI--GKMLGEIV-EPRGPMDFDW 643 ++ L L G + +A+ T IS ++ D K ++ G + GEI+ E RG F + Sbjct: 99 ANMRKLLHELEGKENRKAQFRTAIS-----LILDGKEYLFEGVIKGEIIKEKRGDSGFGY 153 Query: 644 DCVFQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEFKKFL 769 D +F+P + QT+AE+ + KNKIS+R A Q+ +FL Sbjct: 154 DPIFKP----EGYEQTFAELGNETKNKISHRALAVQKLCEFL 191
>sp|Q8U446|HAM1_PYRFU HAM1 protein homolog Length = 185 Score = 69.3 bits (168), Expect = 1e-11 Identities = 48/170 (28%), Positives = 86/170 (50%), Gaps = 2/170 (1%) Frame = +2 Query: 275 IMSQSIDIPEYQEEIIE-ISLLKAKYAYSIIKEPVFVEDTSLCFNALYGMPGPYIKWFVH 451 ++ + ++ PE Q + +E + + + + +P +ED+ L AL G PG Y + Sbjct: 27 VIKKPLEYPEIQADTLEDVVVFGLNWLKDKVDKPFIIEDSGLFIEALNGFPGVYSAYVY- 85 Query: 452 GNLDEKFKKLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFIGKMLGEIV-EPRGP 628 K L G+ K++ G + +A ++I +YD +I F+G++ G I E RG Sbjct: 86 -----KTIGLDGILKLMEGIENRKAYFKSVIGFYDGEI----HLFVGEVRGRISNEKRGL 136 Query: 629 MDFDWDCVFQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEFKKFLLEN 778 F +D +F P +T+AEM + KN +S+R A +EF +++ EN Sbjct: 137 HGFGYDPIFVP----DGFDKTFAEMSTEEKNSVSHRGKALKEFYRWMKEN 182
>sp|Q8PZ91|HAM1_METMA HAM1 protein homolog Length = 184 Score = 68.2 bits (165), Expect = 3e-11 Identities = 52/160 (32%), Positives = 81/160 (50%), Gaps = 2/160 (1%) Frame = +2 Query: 299 PEYQEEIIE-ISLLKAKYAYSIIKEPVFVEDTSLCFNALYGMPGPYIKWFVHGNLDEKFK 475 PE QE+ +E I+ A+Y + + PV V+D+ + NAL G PGPY + FV L Sbjct: 36 PELQEDELEPIAANGAQYVANKLNMPVMVDDSGIFINALNGFPGPYSR-FVEDKLGN--- 91 Query: 476 KLQGLRKMLVGFNDFRAEAVTIISYYDPKIMQDPKAFIGKMLGEIV-EPRGPMDFDWDCV 652 + K++ G D A T+I Y +P Q+P F G + G+I E RG F +D + Sbjct: 92 --PKVLKLMEGEKDRSAYFKTVIGYCEPG--QEPLVFPGVVEGKIAYEERGTGGFGYDPI 147 Query: 653 FQPVAQCQDTLQTYAEMDYKYKNKISNRYNAAQEFKKFLL 772 F+ T+ E+ + KNK+S+R A F ++ + Sbjct: 148 FEYNG------MTFGELGDEEKNKVSHRRRAVDNFLEWFI 181
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 86,541,649
Number of Sequences: 369166
Number of extensions: 1694532
Number of successful extensions: 3761
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3640
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3732
length of database: 68,354,980
effective HSP length: 109
effective length of database: 48,218,865
effective search space used: 8341863645
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail