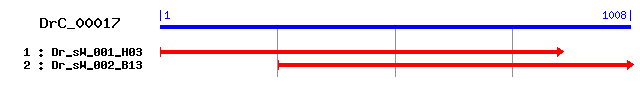
DrC_00017
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00017 (1008 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O14342|YB3C_SCHPO Hypothetical protein C2F12.12c in chro... 121 3e-27 sp|P40480|HOS4_YEAST Protein HOS4 37 0.089 sp|Q61624|ZN148_MOUSE Zinc finger protein 148 (Zinc finger ... 35 0.44 sp|Q62806|ZN148_RAT Zinc finger protein 148 (Zinc finger DN... 34 0.75 sp|Q9UQR1|ZN148_HUMAN Zinc finger protein 148 (Zinc finger ... 33 1.3 sp|Q07283|TRHY_HUMAN Trichohyalin 33 1.3 sp|O22921|WRK25_ARATH Probable WRKY transcription factor 25... 33 1.3 sp|P42349|Y1242_SYNY3 Hypothetical methyltransferase sll124... 32 2.2 sp|Q8BRB7|MYST4_MOUSE Histone acetyltransferase MYST4 (MYST... 32 2.2 sp|Q9H6D3|XKR8_HUMAN XK-related protein 8 32 2.9
>sp|O14342|YB3C_SCHPO Hypothetical protein C2F12.12c in chromosome II Length = 517 Score = 121 bits (304), Expect = 3e-27 Identities = 57/129 (44%), Positives = 81/129 (62%), Gaps = 11/129 (8%) Frame = +3 Query: 606 RKPRFLNRVNTGFDWNKYNQTHYDMDNPPPKIVQGYKFNILYPDLIDKTKTPTFEL---- 773 +KP + NRV GF+WN YNQ H++ +PPPK VQGY+FN+ YPDLI + PT+ + Sbjct: 389 KKPHYFNRVLLGFEWNSYNQAHFNEAHPPPKAVQGYRFNVFYPDLIGTGRAPTYRIERTR 448 Query: 774 ----TPCAD-DDQYAIITFKAGPPYEDIAFKIVNREWETSER--HGYKCQFYRDMLQLYF 932 + D D II F AG PY+DIAF IV+++W+ S + HG+K F +L L+F Sbjct: 449 RKNKSDTTDLQDDVCIIRFIAGEPYQDIAFSIVDKDWDYSAKRDHGFKSSFDNGVLSLHF 508 Query: 933 HFKRYRYRK 959 FK+ +R+ Sbjct: 509 RFKKLHHRR 517
>sp|P40480|HOS4_YEAST Protein HOS4 Length = 1083 Score = 37.0 bits (84), Expect = 0.089 Identities = 50/195 (25%), Positives = 79/195 (40%), Gaps = 4/195 (2%) Frame = +3 Query: 207 KLKHEQGLPVEPLFPSAVVQKEEKVEAKATNSKKTEFILEREEYDHHRYSPVLLSTEEIG 386 KLK L +EP P E + T +++ + E EEY R +E+ Sbjct: 765 KLKSISPLSMEPHSPKKAKSVEISKIHEETAAEREARLKEEEEYRKKRLEKKRKKEQELL 824 Query: 387 DEAIENVIYLAEDDDKKLEFMRKKFLQFGSHKIDQEDELIKLVQEGHEKGESDFNEQFAL 566 + LAED+ K++E K+ K+ + + L K E K E + + Sbjct: 825 QK-------LAEDEKKRIEEQEKQ-------KVLEMERLEKATLEKARKME----REKEM 866 Query: 567 EEQSFLWS--DKYRPRKPRFLNRVNTGFDWNKYNQTHYDMDNPPPKIVQGYKFNILYP-- 734 EE S+ + D Y P + +N N D+ ++ +Y +D K V + IL Sbjct: 867 EEISYRRAVRDLY-PLGLKIIN-FNDKLDYKRFLPLYYFVDEKNDKFVLDLQVMILLKDI 924 Query: 735 DLIDKTKTPTFELTP 779 DL+ K PT E P Sbjct: 925 DLLSKDNQPTSEKIP 939
>sp|Q61624|ZN148_MOUSE Zinc finger protein 148 (Zinc finger DNA binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1) Length = 794 Score = 34.7 bits (78), Expect = 0.44 Identities = 26/98 (26%), Positives = 44/98 (44%), Gaps = 7/98 (7%) Frame = +3 Query: 120 EYFCKLANVLIAKKRLQERHSNNLRLKLIK-----LKHEQGLPVEPLFPSAVVQKEEKVE 284 +YF + VL K+ E H L IK + + G P S +K +K E Sbjct: 262 QYFSRTDRVLKHKRMCHENHDKKLNRCAIKGGLLTSEEDSGFSTSPKDNSLPKKKRQKTE 321 Query: 285 AKATNSKKTEFILEREEY--DHHRYSPVLLSTEEIGDE 392 K++ K E +L++ + D + Y P+ S+ ++ DE Sbjct: 322 KKSSGMDK-ESVLDKSDLKKDKNDYLPLYSSSTKVKDE 358
>sp|Q62806|ZN148_RAT Zinc finger protein 148 (Zinc finger DNA binding protein 89) (Transcription factor ZBP-89) Length = 794 Score = 33.9 bits (76), Expect = 0.75 Identities = 26/98 (26%), Positives = 44/98 (44%), Gaps = 7/98 (7%) Frame = +3 Query: 120 EYFCKLANVLIAKKRLQERHSNNLRLKLIK-----LKHEQGLPVEPLFPSAVVQKEEKVE 284 +YF + VL K+ E H L IK + + G P S +K +K E Sbjct: 262 QYFSRTDRVLKHKRMCHENHDKKLNRCAIKGGLLTSEEDSGFSTSPKDNSLPKKKRQKPE 321 Query: 285 AKATNSKKTEFILEREE--YDHHRYSPVLLSTEEIGDE 392 K++ K E +L++ + D + Y P+ S+ ++ DE Sbjct: 322 KKSSGMDK-ESVLDKSDTKKDRNDYLPLYSSSTKVKDE 358
>sp|Q9UQR1|ZN148_HUMAN Zinc finger protein 148 (Zinc finger DNA binding protein 89) (Transcription factor ZBP-89) Length = 794 Score = 33.1 bits (74), Expect = 1.3 Identities = 26/98 (26%), Positives = 43/98 (43%), Gaps = 7/98 (7%) Frame = +3 Query: 120 EYFCKLANVLIAKKRLQERHSNNLRLKLIK-----LKHEQGLPVEPLFPSAVVQKEEKVE 284 +YF + VL K+ E H L IK + + G P S +K +K E Sbjct: 262 QYFSRTDRVLKHKRMCHENHDKKLNRCAIKGGLLTSEEDSGFSTSPKDNSLPKKKRQKTE 321 Query: 285 AKATNSKKTEFILEREEY--DHHRYSPVLLSTEEIGDE 392 K++ K E L++ + D + Y P+ S+ ++ DE Sbjct: 322 KKSSGMDK-ESALDKSDLKKDKNDYLPLYSSSTKVKDE 358
>sp|Q07283|TRHY_HUMAN Trichohyalin Length = 1898 Score = 33.1 bits (74), Expect = 1.3 Identities = 36/154 (23%), Positives = 70/154 (45%) Frame = +3 Query: 159 KRLQERHSNNLRLKLIKLKHEQGLPVEPLFPSAVVQKEEKVEAKATNSKKTEFILEREEY 338 KR QE + L LK + + +Q L E +EE++E + + LE+EE Sbjct: 587 KREQEERRDQL-LKREEERRQQRLKRE---------QEERLEQRLKREEVER--LEQEER 634 Query: 339 DHHRYSPVLLSTEEIGDEAIENVIYLAEDDDKKLEFMRKKFLQFGSHKIDQEDELIKLVQ 518 R L EE +E ++ E ++++ E +R++ + ++ +E+E +L Q Sbjct: 635 RDER-----LKREEPEEERRHELLKSEEQEERRHEQLRREQQERREQRLKREEEEERLEQ 689 Query: 519 EGHEKGESDFNEQFALEEQSFLWSDKYRPRKPRF 620 + E + EQ EE+ ++ + R P++ Sbjct: 690 RLKREHEEERREQELAEEEQEQARERIKSRIPKW 723
Score = 30.8 bits (68), Expect = 6.4
Identities = 32/142 (22%), Positives = 65/142 (45%), Gaps = 2/142 (1%)
Frame = +3
Query: 156 KKRLQERHSNNLRLKLIKLKHEQGLPVEPLFPSAVVQKEEKVEAKATNSKKTEFIL--ER 329
K+R QER + K ++ K EQ L EP ++E+K + ++ E +L ER
Sbjct: 967 KRRRQERERQYRKDKKLQQKEEQLLGEEP-EKRRRQEREKKYREEEELQQEEEQLLREER 1025
Query: 330 EEYDHHRYSPVLLSTEEIGDEAIENVIYLAEDDDKKLEFMRKKFLQFGSHKIDQEDELIK 509
E+ + +E+ E E + E + ++L+ +++ + ++ QE+E +
Sbjct: 1026 EKRRRQEWERQYRKKDELQQE--EEQLLREEREKRRLQERERQYRE--EEELQQEEEQL- 1080
Query: 510 LVQEGHEKGESDFNEQFALEEQ 575
L +E + + Q+ EE+
Sbjct: 1081 LGEERETRRRQELERQYRKEEE 1102
>sp|O22921|WRK25_ARATH Probable WRKY transcription factor 25 (WRKY DNA-binding protein 25) Length = 393 Score = 33.1 bits (74), Expect = 1.3 Identities = 23/104 (22%), Positives = 39/104 (37%), Gaps = 2/104 (1%) Frame = +3 Query: 519 EGHEKGESDFNEQFALEEQSFLWSDKYRPRKPRFL--NRVNTGFDWNKYNQTHYDMDNPP 692 + HEK + + A + + + + + + P ++ N G+ W KY Q P Sbjct: 126 QDHEKKQEMIPNEIATQNNNQSFGTERQIKIPAYMVSRNSNDGYGWRKYGQKQVKKSENP 185 Query: 693 PKIVQGYKFNILYPDLIDKTKTPTFELTPCADDDQYAIITFKAG 824 F YPD + K ++ A D Q I +K G Sbjct: 186 RSY-----FKCTYPDCVSK------KIVETASDGQITEIIYKGG 218
>sp|P42349|Y1242_SYNY3 Hypothetical methyltransferase sll1242 (ORF N) Length = 536 Score = 32.3 bits (72), Expect = 2.2 Identities = 17/45 (37%), Positives = 24/45 (53%) Frame = +3 Query: 792 DQYAIITFKAGPPYEDIAFKIVNREWETSERHGYKCQFYRDMLQL 926 +Q ++ F P EDIA + V WE + HGY + YR L+L Sbjct: 380 NQTTLMNFIPTRPLEDIANEYVEAFWELYDPHGYLDRNYRCFLKL 424
>sp|Q8BRB7|MYST4_MOUSE Histone acetyltransferase MYST4 (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein) Length = 1872 Score = 32.3 bits (72), Expect = 2.2 Identities = 24/112 (21%), Positives = 48/112 (42%), Gaps = 1/112 (0%) Frame = +3 Query: 213 KHEQGLPVEP-LFPSAVVQKEEKVEAKATNSKKTEFILEREEYDHHRYSPVLLSTEEIGD 389 + E G+PV P P V +E+ + + ++ E EREE + EE+ Sbjct: 1128 REETGIPVSPHKSPGGKVDEEDLIRGEEEGEEEGEEEGEREEQEEE---------EEVTT 1178 Query: 390 EAIENVIYLAEDDDKKLEFMRKKFLQFGSHKIDQEDELIKLVQEGHEKGESD 545 E + E+ + ++ ++ + G H+ D+++E E H+ + D Sbjct: 1179 EKDLDGAKSKENPEPEISMEKEDPVHLGDHEEDEDEEEEPSHNEDHDADDED 1230
>sp|Q9H6D3|XKR8_HUMAN XK-related protein 8 Length = 395 Score = 32.0 bits (71), Expect = 2.9 Identities = 16/38 (42%), Positives = 21/38 (55%) Frame = -1 Query: 435 FYHHLPLNKSHFRWLHRRFLQWITKPDCIDGDRTLLSP 322 F+ L L ++ WLH W PD +DG R+LLSP Sbjct: 327 FFLGLALRLVYYHWLHPSCC-WKPDPDQVDGARSLLSP 363
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 107,858,743
Number of Sequences: 369166
Number of extensions: 2204264
Number of successful extensions: 6380
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6122
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6374
length of database: 68,354,980
effective HSP length: 111
effective length of database: 47,849,395
effective search space used: 10718264480
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail