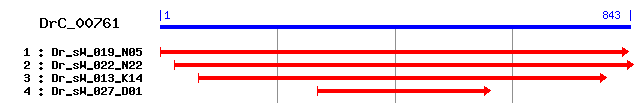
DrC_00761
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00761 (843 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O55101|SNG2_MOUSE Synaptogyrin-2 (Cellugyrin) 42 0.002 sp|O54980|SNG2_RAT Synaptogyrin-2 (Cellugyrin) 40 0.008 sp|O95473|SNG4_HUMAN Synaptogyrin-4 39 0.014 sp|O43760|SNG2_HUMAN Synaptogyrin-2 (Cellugyrin) 39 0.018 sp|Q9Z1L2|SNG4_MOUSE Synaptogyrin-4 38 0.040 sp|O43759|SNG1_HUMAN Synaptogyrin-1 37 0.089 sp|O55100|SNG1_MOUSE Synaptogyrin-1 35 0.26 sp|Q62876|SNG1_RAT Synaptogyrin-1 (p29) 35 0.26 sp|P16537|SPC3_STRPU SPEC3 protein 33 0.99 sp|P87293|YDN1_SCHPO Hypothetical protein C16A10.01 in chro... 31 3.7
>sp|O55101|SNG2_MOUSE Synaptogyrin-2 (Cellugyrin) Length = 224 Score = 42.0 bits (97), Expect = 0.002 Identities = 22/93 (23%), Positives = 42/93 (45%), Gaps = 1/93 (1%) Frame = +1 Query: 169 FCVFNMQTPSCDYAQSVGWISIIFGIYVIILE-IFSQSINLKFLKISFIFEFLINVILSI 345 +CVFN +C Y ++G ++ + + ++++ FSQ N K I + L + + + Sbjct: 58 YCVFNQNEDACRYGSAIGVLAFLASAFFLVVDAFFSQISNATDRKYLVIGDLLFSALWTF 117 Query: 346 MWLAAFIDLNVKWTNVPNMPESWVLANARAVIS 444 +W F L +W +ARA I+ Sbjct: 118 LWFVGFCFLTNQWAATKPQDVRVGADSARAAIT 150
>sp|O54980|SNG2_RAT Synaptogyrin-2 (Cellugyrin) Length = 224 Score = 40.0 bits (92), Expect = 0.008 Identities = 18/72 (25%), Positives = 35/72 (48%), Gaps = 1/72 (1%) Frame = +1 Query: 172 CVFNMQTPSCDYAQSVGWISIIFGIYVIILE-IFSQSINLKFLKISFIFEFLINVILSIM 348 CVFN +C Y ++G ++ + + ++++ FSQ N K I + L + + + + Sbjct: 59 CVFNRNEDACRYGSAIGVLAFLASAFFLVVDAFFSQISNATDRKYLVIGDLLFSALWTFL 118 Query: 349 WLAAFIDLNVKW 384 W F L +W Sbjct: 119 WFVGFCFLTNQW 130
>sp|O95473|SNG4_HUMAN Synaptogyrin-4 Length = 234 Score = 39.3 bits (90), Expect = 0.014 Identities = 19/89 (21%), Positives = 47/89 (52%), Gaps = 1/89 (1%) Frame = +1 Query: 172 CVFNMQTPSCDYAQSVGWISIIFGIYVIILEIFSQSINLKFLKISF-IFEFLINVILSIM 348 C+ N + +C +A G+++ + + ++L+ I K +F + +F++ V+ +++ Sbjct: 57 CILNSNSVACSFAVGAGFLAFLSCLAFLVLDTQETRIAGTRFKTAFQLLDFILAVLWAVV 116 Query: 349 WLAAFIDLNVKWTNVPNMPESWVLANARA 435 W F L +W + P P+ ++L ++ A Sbjct: 117 WFMGFCFLANQWQHSP--PKEFLLGSSSA 143
>sp|O43760|SNG2_HUMAN Synaptogyrin-2 (Cellugyrin) Length = 224 Score = 38.9 bits (89), Expect = 0.018 Identities = 17/73 (23%), Positives = 35/73 (47%), Gaps = 1/73 (1%) Frame = +1 Query: 169 FCVFNMQTPSCDYAQSVGWISIIFGIYVIILE-IFSQSINLKFLKISFIFEFLINVILSI 345 +CVFN +C Y ++G ++ + + ++++ F Q N K I + L + + + Sbjct: 58 YCVFNRNEDACRYGSAIGVLAFLASAFFLVVDAYFPQISNATDRKYLVIGDLLFSALWTF 117 Query: 346 MWLAAFIDLNVKW 384 +W F L +W Sbjct: 118 LWFVGFCFLTNQW 130
>sp|Q9Z1L2|SNG4_MOUSE Synaptogyrin-4 Length = 233 Score = 37.7 bits (86), Expect = 0.040 Identities = 20/89 (22%), Positives = 47/89 (52%), Gaps = 1/89 (1%) Frame = +1 Query: 172 CVFNMQTPSCDYAQSVGWISIIFGIYVIILEIFSQSINLKFLKISF-IFEFLINVILSIM 348 CV N +C +A G++S + + + ++ + + + KI+F + +F++ V+ + + Sbjct: 57 CVLNSNHMACSFAVGAGFLSFLSCLVFLAIDAYERRLVGTRFKIAFQLLDFILAVLWAGV 116 Query: 349 WLAAFIDLNVKWTNVPNMPESWVLANARA 435 W AF L +W + + + ++L N+ A Sbjct: 117 WFVAFCFLASQWQH--SKSKHFLLGNSSA 143
>sp|O43759|SNG1_HUMAN Synaptogyrin-1 Length = 234 Score = 36.6 bits (83), Expect = 0.089 Identities = 42/190 (22%), Positives = 76/190 (40%), Gaps = 9/190 (4%) Frame = +1 Query: 169 FCVFNMQTPSCDYAQSVGWISIIFGIYVIILEI-FSQSINLKFLKISFIFEFLINVILSI 345 FC++N +C Y +VG ++ + + + L++ F Q ++K K + + + ++ + Sbjct: 58 FCIYNRNPNACSYGVAVGVLAFLTCLLYLALDVYFPQISSVKDRKKAVLSDIGVSAFWAF 117 Query: 346 MWLAAFIDLNVKW--TNVPNMPESWVLANARAVISXXXXXXXIQVLLVISVICGLIQIIR 519 +W F L +W + + P + ARA I+ S+ Q + Sbjct: 118 LWFVGFCYLANQWQVSKPKDNPLNEGTDAARAAIA----------FSFFSIFTWAGQAVL 167 Query: 520 AMGQGDPTAEQPVKDNDNNNIDQTYRLKFTLTRFYYVVSNTGVAVV----QHQQEVNTFH 687 A + A+ + D + Q + + YV NTG +QQ NTF Sbjct: 168 AFQRYQIGADSALFSQDYMDPSQDSSMPYA----PYVEPNTGPDPAGMGGTYQQPANTFD 223 Query: 688 T--SSYQSIG 711 T YQS G Sbjct: 224 TEPQGYQSQG 233
>sp|O55100|SNG1_MOUSE Synaptogyrin-1 Length = 234 Score = 35.0 bits (79), Expect = 0.26 Identities = 16/73 (21%), Positives = 36/73 (49%), Gaps = 1/73 (1%) Frame = +1 Query: 169 FCVFNMQTPSCDYAQSVGWISIIFGIYVIILEI-FSQSINLKFLKISFIFEFLINVILSI 345 FC++N +C Y +VG ++ + + + L++ F Q ++K K + + + ++ + Sbjct: 58 FCIYNRNPNACSYGVTVGVLAFLTCLLYLALDVYFPQISSVKDRKKAVLSDIGVSAFWAF 117 Query: 346 MWLAAFIDLNVKW 384 W F L +W Sbjct: 118 FWFVGFCFLANQW 130
>sp|Q62876|SNG1_RAT Synaptogyrin-1 (p29) Length = 234 Score = 35.0 bits (79), Expect = 0.26 Identities = 16/73 (21%), Positives = 36/73 (49%), Gaps = 1/73 (1%) Frame = +1 Query: 169 FCVFNMQTPSCDYAQSVGWISIIFGIYVIILEI-FSQSINLKFLKISFIFEFLINVILSI 345 FC++N +C Y +VG ++ + + + L++ F Q ++K K + + + ++ + Sbjct: 58 FCIYNRNPNACSYGVTVGVLAFLTCLVYLALDVYFPQISSVKDRKKAVLSDIGVSAFWAF 117 Query: 346 MWLAAFIDLNVKW 384 W F L +W Sbjct: 118 FWFVGFCFLANQW 130
>sp|P16537|SPC3_STRPU SPEC3 protein Length = 208 Score = 33.1 bits (74), Expect = 0.99 Identities = 23/77 (29%), Positives = 34/77 (44%) Frame = +1 Query: 154 SFLPGFCVFNMQTPSCDYAQSVGWISIIFGIYVIILEIFSQSINLKFLKISFIFEFLINV 333 + + GF VF P D VG + I F I + L+I ++ F + Sbjct: 125 TIIAGFAVFCCGNPGADGGSKVGTMCINFWIGL--------------LQIGTVWFFFLGW 170 Query: 334 ILSIMWLAAFIDLNVKW 384 I SIMW AAFI ++ + Sbjct: 171 IWSIMWGAAFIGMSADY 187
>sp|P87293|YDN1_SCHPO Hypothetical protein C16A10.01 in chromosome I Length = 830 Score = 31.2 bits (69), Expect = 3.7 Identities = 15/44 (34%), Positives = 26/44 (59%), Gaps = 1/44 (2%) Frame = +1 Query: 220 GWISIIFGIYV-IILEIFSQSINLKFLKISFIFEFLINVILSIM 348 GW ++ G + ++L IF IN +F +FE +I++ILS + Sbjct: 526 GWRDMLIGFVLGLLLGIFRVYINPRFFLFDSLFEVIISIILSFL 569
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 88,263,400
Number of Sequences: 369166
Number of extensions: 1688625
Number of successful extensions: 3819
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3676
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3815
length of database: 68,354,980
effective HSP length: 109
effective length of database: 48,218,865
effective search space used: 8245425915
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail