DrC_00757
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00757 (916 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9W3C2|GSCR2_DROME Hypothetical protein CG1785 41 0.005 sp|P14873|MAP1B_MOUSE Microtubule-associated protein 1B (MA... 36 0.13 sp|P53935|YNJ1_YEAST Hypothetical 141.5 kDa protein in YPT5... 35 0.38 sp|Q9NEU5|GSCR2_CAEEL Hypothetical protein Y39B6A.33 in chr... 34 0.50 sp|Q14152|IF3A_HUMAN Eukaryotic translation initiation fact... 34 0.50 sp|Q28820|TRDN_RABIT Triadin 34 0.50 sp|Q8CG47|SMC4_MOUSE Structural maintenance of chromosomes ... 34 0.65 sp|Q9MTH5|YCF1_OENHO Hypothetical 287.9 kDa protein ycf1 33 0.85 sp|O14617|AP3D1_HUMAN Adapter-related protein complex 3 del... 33 0.85 sp|Q60YI3|NOLA2_CAEBR Putative H/ACA ribonucleoprotein comp... 33 0.85
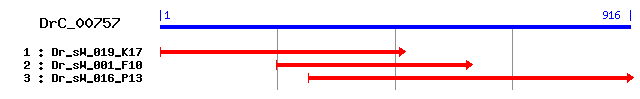
>sp|Q9W3C2|GSCR2_DROME Hypothetical protein CG1785 Length = 478 Score = 40.8 bits (94), Expect = 0.005 Identities = 52/258 (20%), Positives = 99/258 (38%), Gaps = 45/258 (17%) Frame = +3 Query: 195 KQKSAMEHKSKHAKPSEIIHPGTSWFPVKSEHNELLE--------MLEKDETKRIIEDKK 350 K +++ HK+ AK E+ HPG S+ P +H L++ +++K++ + + Sbjct: 195 KTHASLHHKTTKAKKFELPHPGMSYNPAPEDHQALIDQVVEREEGIIKKEQHLKRVTTSM 254 Query: 351 FTFKTVEE------IEMKXXXXXXXXXXXXXXXSTRISAKK----------NKPLTSKER 482 F+ T EE EM T K+ N P+ +K++ Sbjct: 255 FSKVTPEERDRRRLNEMSQGMDEEEGSELDEDVQTNGKKKEDDDEKPYHTINAPVENKKK 314 Query: 483 KRKLMKMNLEMNKQKERSKQ-----------------KGEAHELRNIPKLCNEIKRTXXX 611 ++ + L KQKE ++Q K HEL + + N++K+ Sbjct: 315 SKQARRNEL---KQKELARQTELKRKLKQQTADLIRIKSIRHELDDEEEDLNDLKKRRKQ 371 Query: 612 XXXXXXXXXXXXXXXXXDKIFREQIHRMPFQLPSELAPNLRSMQTEGDLLQELCDKVN-- 785 ++ + LP ++A NLR+++TE LL++ + Sbjct: 372 RAEKAKFEPKRLGRHKFEE------PDLDVNLPEDIAGNLRNVKTESSLLKDRFHTLQRN 425 Query: 786 --LHRYKMPNRKNEKFEK 833 L K+ +RK K ++ Sbjct: 426 NMLPTTKLVSRKKSKVKR 443
>sp|P14873|MAP1B_MOUSE Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1] Length = 2464 Score = 36.2 bits (82), Expect = 0.13 Identities = 51/210 (24%), Positives = 91/210 (43%), Gaps = 10/210 (4%) Frame = +3 Query: 3 DIIKSNQLREIDEA-SFEVTNKLKSKELKYSTVPRIITKKI-----RKFVKEEVIDLWAK 164 ++ K++Q+ + + S E K K +K + P + K++ + VK EV + A Sbjct: 567 EVTKTSQVEKTPKVESKEKVLVKKDKPVKTESKPSVTEKEVSSKEEQSPVKAEVAEKQAT 626 Query: 165 DSNDT----QVLSKKQKSAMEHKSKHAKPSEIIHPGTSWFPVKSEHNELLEMLEKDETKR 332 +S +V+ K+ K+ +E K K KP + + VK E L+KDE R Sbjct: 627 ESKPKVTKDKVVKKEIKTKLEEK-KEEKPKKEV--------VKKEDK---TPLKKDEKPR 674 Query: 333 IIEDKKFTFKTVEEIEMKXXXXXXXXXXXXXXXSTRISAKKNKPLTSKERKRKLMKMNLE 512 E KK K +++ E K + KK PL K+ K+ E Sbjct: 675 KEEVKKEIKKEIKKEERK---------------ELKKEVKKETPL--KDAKK-------E 710 Query: 513 MNKQKERSKQKGEAHELRNIPKLCNEIKRT 602 + K++++ +K E + I K+ +IK++ Sbjct: 711 VKKEEKKEVKKEEKEPKKEIKKISKDIKKS 740
>sp|P53935|YNJ1_YEAST Hypothetical 141.5 kDa protein in YPT53-RHO2 intergenic region Length = 1240 Score = 34.7 bits (78), Expect = 0.38 Identities = 34/134 (25%), Positives = 56/134 (41%), Gaps = 1/134 (0%) Frame = +3 Query: 180 QVLSKKQKSAMEHKSKHAKPSEIIHPGTSWFPVKSEHNELLEMLEKDETKRIIEDKKFTF 359 Q L ++++ E + K K E K + + L+ L K+E KR E++K Sbjct: 629 QELEEEKRKKREKEEKKQKKRE-----------KEKEKKRLQQLAKEEEKRKREEEKERL 677 Query: 360 KT-VEEIEMKXXXXXXXXXXXXXXXSTRISAKKNKPLTSKERKRKLMKMNLEMNKQKERS 536 K +EE EM+ + R ++ K +ERKR+L + Q+++ Sbjct: 678 KKELEEREMRRRE------------AQRKKVEEAKRKKDEERKRRLEEQQRREEMQEKQR 725 Query: 537 KQKGEAHELRNIPK 578 KQK E R K Sbjct: 726 KQKEELKRKREEEK 739
>sp|Q9NEU5|GSCR2_CAEEL Hypothetical protein Y39B6A.33 in chromosome V Length = 438 Score = 34.3 bits (77), Expect = 0.50 Identities = 48/245 (19%), Positives = 93/245 (37%), Gaps = 17/245 (6%) Frame = +3 Query: 186 LSKKQKSAMEHKSKHAKPSEIIHPGTSWFPVKSEHNELLEMLE----------KDETKRI 335 ++ +++ ++H++K E W V +EH LEM E DE + Sbjct: 205 IAGEEQKLIDHEAKIKAGIE-----PQWEKVTTEHERFLEMAEGLRIHPKYGKDDEEEEE 259 Query: 336 IEDKKFTFKTVEEIEMKXXXXXXXXXXXXXXXSTRISAKKNKPLTSKERKRKLMKMNLEM 515 + + + KT E E K S R+ + K++K K K++ E Sbjct: 260 AGNSEKSMKTGGEAEPK---------------SQRVECDR-MTKEQKKKKAKAQKLDKEE 303 Query: 516 NKQKERSKQKGEAHELRNIPKLCNEIKRTXXXXXXXXXXXXXXXXXXXXDKIFREQIHR- 692 ++ E ++ ++H + +L E+ K R+Q+ + Sbjct: 304 KRRLEEKAKEQDSHNVYRTKQLHKELDEEEKQRHEESEVRKKEKLINKLTK--RQQLGKG 361 Query: 693 ------MPFQLPSELAPNLRSMQTEGDLLQELCDKVNLHRYKMPNRKNEKFEKMIKREAS 854 PF L EL NLR ++ +G +L + +L R M +K ++ IK Sbjct: 362 KFVDAEDPFLLQEELTGNLRQLKPQGHVLDDRMK--SLQRRNMLPIGGDKEKRRIKNRLK 419 Query: 855 FELIK 869 ++++ Sbjct: 420 SKVVE 424
>sp|Q14152|IF3A_HUMAN Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) Length = 1382 Score = 34.3 bits (77), Expect = 0.50 Identities = 39/196 (19%), Positives = 82/196 (41%), Gaps = 8/196 (4%) Frame = +3 Query: 3 DIIKSNQLREIDEASFEVTNKLKSKELK---YSTVPRIITKKIRKFVKEEV----IDLWA 161 D I + Q+ ++++ E+ +LK++E K + R+ + K EE +DLW Sbjct: 667 DFIMAKQVEQLEKEKKELQERLKNQEKKIDYFERAKRLEEIPLIKSAYEEQRIKDMDLWE 726 Query: 162 KDSNDT-QVLSKKQKSAMEHKSKHAKPSEIIHPGTSWFPVKSEHNELLEMLEKDETKRII 338 + + + +++ A+EHK++ ++ E +L M K + + Sbjct: 727 QQEEERITTMQLEREKALEHKNRMSR--------------MLEDRDLFVMRLKAARQSVY 772 Query: 339 EDKKFTFKTVEEIEMKXXXXXXXXXXXXXXXSTRISAKKNKPLTSKERKRKLMKMNLEMN 518 E+K K EE + RI+ + K +E +R+ + L+ Sbjct: 773 EEK---LKQFEERLAEERHNRLEERKRQRKEERRITYYREK---EEEEQRRAEEQMLKER 826 Query: 519 KQKERSKQKGEAHELR 566 +++ER+++ ELR Sbjct: 827 EERERAERAKREEELR 842
>sp|Q28820|TRDN_RABIT Triadin Length = 706 Score = 34.3 bits (77), Expect = 0.50 Identities = 36/129 (27%), Positives = 58/129 (44%), Gaps = 1/129 (0%) Frame = +3 Query: 3 DIIKSNQLREIDEASFEVTN-KLKSKELKYSTVPRIITKKIRKFVKEEVIDLWAKDSNDT 179 DII S+ E DE + +++ LK + + +KI K K E T Sbjct: 112 DIISSDGDEEDDEGDEDTAKGEIEEPPLKRKDIHK---EKIEKQEKPE-------RKIPT 161 Query: 180 QVLSKKQKSAMEHKSKHAKPSEIIHPGTSWFPVKSEHNELLEMLEKDETKRIIEDKKFTF 359 +V+ K+++ E + KP + K+ H E LE EK ETK + +++K Sbjct: 162 KVVHKEKEKEKEKVKEKEKPEK-----------KATHKEKLEKKEKPETKTVTKEEK-KA 209 Query: 360 KTVEEIEMK 386 +T E+IE K Sbjct: 210 RTKEKIEEK 218
>sp|Q8CG47|SMC4_MOUSE Structural maintenance of chromosomes 4-like 1 protein (Chromosome-associated polypeptide C) (XCAP-C homolog) Length = 1286 Score = 33.9 bits (76), Expect = 0.65 Identities = 53/233 (22%), Positives = 96/233 (41%), Gaps = 44/233 (18%) Frame = +3 Query: 27 REIDEASFEVTNKLKSKELKYSTVPRIITKKIRKFVKEEVIDLWAKDSNDTQV------- 185 +EI E S ++N++K+K V + + K + KF+++ D D QV Sbjct: 347 KEITEKSNVLSNEMKAKNSAVKDVEKKLNK-VTKFIEQNKEKFTQLDLEDVQVREKLKHA 405 Query: 186 ----------LSKKQKSAMEHKSKHAKPSEIIHPGTSWFPV-----KSEHNELLEMLE-- 314 L K ++ E KS AK +I+ T+ + E +L E+++ Sbjct: 406 TSKAKKLEKQLQKDKEKVEELKSVPAKSKTVINETTTRNNSLEKEREKEEKKLKEVMDSL 465 Query: 315 KDETKRIIEDKKFTFK-------TVEEIEMKXXXXXXXXXXXXXXXSTRIS--AKKNKPL 467 K ET+ + ++K+ K +V E K +T +S +K + L Sbjct: 466 KQETQGLQKEKEIQEKELMGFNKSVNEARSKMEVAQSELDIYLSRHNTAVSQLSKAKEAL 525 Query: 468 -----TSKERKRKLMKMNLEMN------KQKERSKQKGEAHELRNIPKLCNEI 593 T KERK + +N ++ K+KE+ QK E+ N+ L +++ Sbjct: 526 ITASETLKERKAAIKDINTKLPQTQQELKEKEKELQKLTQEEI-NLKSLVHDL 577
>sp|Q9MTH5|YCF1_OENHO Hypothetical 287.9 kDa protein ycf1 Length = 2434 Score = 33.5 bits (75), Expect = 0.85 Identities = 17/54 (31%), Positives = 30/54 (55%) Frame = +3 Query: 438 RISAKKNKPLTSKERKRKLMKMNLEMNKQKERSKQKGEAHELRNIPKLCNEIKR 599 +IS K K + SK++ K + N++ ++ +SKQ ++ I + NEIKR Sbjct: 770 KISKSKQKNVKSKQKNVKSKQKNVKSKQKNVKSKQNEIKRKVNEIKRKVNEIKR 823
Score = 32.3 bits (72), Expect = 1.9
Identities = 17/53 (32%), Positives = 28/53 (52%)
Frame = +3
Query: 441 ISAKKNKPLTSKERKRKLMKMNLEMNKQKERSKQKGEAHELRNIPKLCNEIKR 599
+ KK K SK++ K + N++ ++ +SKQK + I + NEIKR
Sbjct: 764 VPKKKKKISKSKQKNVKSKQKNVKSKQKNVKSKQKNVKSKQNEIKRKVNEIKR 816
>sp|O14617|AP3D1_HUMAN Adapter-related protein complex 3 delta 1 subunit (Delta-adaptin 3) (AP-3 complex delta subunit) (Delta-adaptin) Length = 1153 Score = 33.5 bits (75), Expect = 0.85 Identities = 33/139 (23%), Positives = 53/139 (38%) Frame = +3 Query: 162 KDSNDTQVLSKKQKSAMEHKSKHAKPSEIIHPGTSWFPVKSEHNELLEMLEKDETKRIIE 341 KD + K++K H S + E I P V E E L DE + Sbjct: 739 KDKRRKKRKEKEKKGKRRHSSLPTESDEDIAPAQQVDIVTEEMPE--NALPSDEDDK--- 793 Query: 342 DKKFTFKTVEEIEMKXXXXXXXXXXXXXXXSTRISAKKNKPLTSKERKRKLMKMNLEMNK 521 D ++ ++ I++ +T S K + E+K K K + +K Sbjct: 794 DPNDPYRALD-IDLDKPLADSEKLPIQKHRNTETSKSPEKDVPMVEKKSKKPKKKEKKHK 852 Query: 522 QKERSKQKGEAHELRNIPK 578 +KER K+K + E + PK Sbjct: 853 EKERDKEKKKEKEKKKSPK 871
>sp|Q60YI3|NOLA2_CAEBR Putative H/ACA ribonucleoprotein complex subunit 2-like protein Length = 163 Score = 33.5 bits (75), Expect = 0.85 Identities = 20/51 (39%), Positives = 31/51 (60%), Gaps = 1/51 (1%) Frame = +3 Query: 711 SELAPNLRSMQTEGDLLQELCDKVNLHRYKMPNRK-NEKFEKMIKREASFE 860 SE+A ++ ++ TE D Q LC+ VN + NRK +K K+IK+ A+ E Sbjct: 16 SEIAEDVAALTTEKDEYQALCELVNPIAQPLANRKLAKKVYKLIKKAAAGE 66
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 84,185,906
Number of Sequences: 369166
Number of extensions: 1540211
Number of successful extensions: 5750
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5394
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5724
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 9318621220
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail