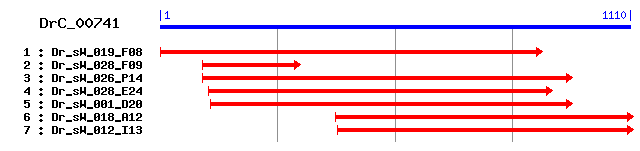
DrC_00741
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00741 (1110 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P04929|HRPX_PLALO Histidine-rich glycoprotein precursor 58 4e-08 sp|Q9V3A4|CSUP_DROME Catecholamines up protein 49 3e-05 sp|P05227|HRP1_PLAFA Histidine-rich protein precursor (Clon... 48 4e-05 sp|P08723|SPBP_RAT Prostatic spermine-binding protein precu... 46 2e-04 sp|Q92504|KE4_HUMAN Zinc transporter SLC39A7 (Solute carrie... 45 3e-04 sp|P04196|HRG_HUMAN Histidine-rich glycoprotein precursor (... 42 0.002 sp|Q31125|KE4_MOUSE Zinc transporter SLC39A7 (Solute carrie... 41 0.005 sp|P08016|EGGS_SCHMA Putative eggshell protein 40 0.012 sp|O67204|YB27_AQUAE Hypothetical protein AQ_1127 40 0.016 sp|Q9XTQ7|KE4L_CAEEL Hypothetical Ke4-like protein H13N06.5... 38 0.046
>sp|P04929|HRPX_PLALO Histidine-rich glycoprotein precursor Length = 351 Score = 58.2 bits (139), Expect = 4e-08 Identities = 28/93 (30%), Positives = 38/93 (40%), Gaps = 3/93 (3%) Frame = -3 Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVD---NVHDPHDVYDVHGPHDED 572 ++H H D +H +D +HH D + H D + H HD + H HD Sbjct: 255 HHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHDAH 314 Query: 571 CVHGPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473 H H + H HH D H H+ D H HH Sbjct: 315 HHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHH 347
Score = 56.2 bits (134), Expect = 2e-07
Identities = 29/93 (31%), Positives = 39/93 (41%), Gaps = 1/93 (1%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVH 563
++H H D +H +D +HH D + H D H H +D H H H
Sbjct: 245 HHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHH-----H 299
Query: 562 GPHDGDNVHGHHD-EDCVHGHYDAEDVHGHHDE 467
HD + H HHD H H+DA H HH +
Sbjct: 300 HHHDAHHHHHHHDAHHHHHHHHDAHHHHHHHHD 332
Score = 55.5 bits (132), Expect = 3e-07
Identities = 27/93 (29%), Positives = 37/93 (39%), Gaps = 3/93 (3%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVD---NVHDPHDVYDVHGPHDED 572
++H H +H +D +HH D + H D + H HD + H H +
Sbjct: 235 HHHHHHHHGHHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDA 294
Query: 571 CVHGPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
H H D H HH D H H+ D H HH
Sbjct: 295 HHHHHHHHDAHHHHHHHDAHHHHHHHHDAHHHH 327
Score = 51.2 bits (121), Expect = 5e-06
Identities = 26/90 (28%), Positives = 35/90 (38%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVH 563
++H H +H + +HH D + H D H H +D H H H
Sbjct: 225 HHHHHHHHGHHHHHHHHHGHHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHH-----H 279
Query: 562 GPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
HD + H HH D H H+ D H HH
Sbjct: 280 HHHDAHH-HHHHHHDAHHHHHHHHDAHHHH 308
Score = 45.1 bits (105), Expect = 4e-04
Identities = 26/92 (28%), Positives = 34/92 (36%), Gaps = 1/92 (1%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDG-DIYDHSDVDNVHDPHDVYDVHGPHDEDCV 566
++H H +H +HHD + H D + H H H H D
Sbjct: 226 HHHHHHHGHHHHHHHHHGHHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAH 285
Query: 565 HGPHDGDNVHGHHDEDCVHGHYDAEDVHGHHD 470
H H + H HH H H+DA H HHD
Sbjct: 286 HHHHHHHDAHHHH-----HHHHDAHHHHHHHD 312
Score = 44.3 bits (103), Expect = 6e-04
Identities = 24/90 (26%), Positives = 34/90 (37%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVH 563
++H H +H + +HH + H + H H +D H H H
Sbjct: 205 HHHHHHHHAPHHHHHHHHGHHHHHHHHHGHHHHHHHHHGHHHHHHHHHDAHHHH-----H 259
Query: 562 GPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
HD + H HH D H H+ D H HH
Sbjct: 260 HHHDAHHHH-HHHHDAHHHHHHHHDAHHHH 288
Score = 43.9 bits (102), Expect = 8e-04
Identities = 24/92 (26%), Positives = 34/92 (36%), Gaps = 2/92 (2%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVH 563
++H H +H + +HH + H + H HD + H H D H
Sbjct: 210 HHHAPHHHHHHHHGHHHHHHHHHGHHHHHH--HHHGHHHHHHHHHDAHH-HHHHHHDAHH 266
Query: 562 GPHDGDNV--HGHHDEDCVHGHYDAEDVHGHH 473
H + H HH D H H+ D H HH
Sbjct: 267 HHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHH 298
Score = 43.5 bits (101), Expect = 0.001
Identities = 24/89 (26%), Positives = 32/89 (35%)
Frame = -3
Query: 739 YHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHG 560
+H H +H + +HH G + H H H + H H H
Sbjct: 195 HHHHHHHHAPHHHHHHHHAPHHHHHHHHGHHHHHHHHHGHHHHHHHHHGHHHHH----HH 250
Query: 559 PHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
HD + H HH D H H+ D H HH
Sbjct: 251 HHDAHHHH-HHHHDAHHHHHHHHDAHHHH 278
Score = 42.7 bits (99), Expect = 0.002
Identities = 24/90 (26%), Positives = 32/90 (35%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVH 563
++H H +H +HH + H + H H HG H H
Sbjct: 186 HHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHGHHHHHHHH----HGHHHH---H 238
Query: 562 GPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
H G + H HH D H H+ D H HH
Sbjct: 239 HHHHGHHHHHHHHHDAHHHHHHHHDAHHHH 268
Score = 42.7 bits (99), Expect = 0.002
Identities = 23/92 (25%), Positives = 34/92 (36%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVH 563
++H H +H + +HH + H + H H + H H D H
Sbjct: 200 HHHAPHHHHHHHHAPHHHHHHHHGHHHHH---HHHHGHHHHHHHHHGHHHHHHHHHDAHH 256
Query: 562 GPHDGDNVHGHHDEDCVHGHYDAEDVHGHHDE 467
H + H HH H H+DA H HH +
Sbjct: 257 HHHHHHDAHHHH-----HHHHDAHHHHHHHHD 283
Score = 41.6 bits (96), Expect = 0.004
Identities = 32/126 (25%), Positives = 53/126 (42%), Gaps = 3/126 (2%)
Frame = -3
Query: 835 SVSLLRHRLKMFHNLSK**ILISKIYNAIIFYYHIDFHDDFENHDDQKYDIQDQSNHHDD 656
S SL++H + NL+ +L+ + + H E+H+ ++ +HH +
Sbjct: 27 SSSLVKHIPQTGSNLTFDRVLVEDTVHPEHLHEEHHHHHPEEHHEPHH---EEHHHHHPE 83
Query: 655 GDIYDHSDVDNVHDPHDVYD--VHGPHDEDCV-HGPHDGDNVHGHHDEDCVHGHYDAEDV 485
H + + H PH + H PH + H H H HH E+ H H+ A
Sbjct: 84 EHHEPHHEEHHHHHPHPHHHHHHHPPHHHHHLGHHHHHHHAAHHHHHEEHHHHHHAAH-- 141
Query: 484 HGHHDE 467
H HH+E
Sbjct: 142 HHHHEE 147
Score = 40.8 bits (94), Expect = 0.007
Identities = 24/94 (25%), Positives = 33/94 (35%), Gaps = 3/94 (3%)
Frame = -3
Query: 739 YHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHG 560
+H H +H + +HH + H H H + H H H
Sbjct: 185 HHHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHGHHHHHHHHHGHHHH-----HH 239
Query: 559 PHDGDNVHGHHDEDCVHGHY---DAEDVHGHHDE 467
H G + H HH D H H+ DA H HH +
Sbjct: 240 HHHGHHHHHHHHHDAHHHHHHHHDAHHHHHHHHD 273
Score = 40.0 bits (92), Expect = 0.012
Identities = 23/98 (23%), Positives = 34/98 (34%), Gaps = 8/98 (8%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNV--------HDPHDVYDVHG 587
++H+ +H H + +HH + H + H PH + H
Sbjct: 163 HHHLGYHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHH 222
Query: 586 PHDEDCVHGPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
H H H G + H HH H H+ D H HH
Sbjct: 223 GHHHH--HHHHHGHHHHHHHHHGHHHHHHHHHDAHHHH 258
Score = 38.5 bits (88), Expect = 0.035
Identities = 22/91 (24%), Positives = 31/91 (34%)
Frame = -3
Query: 739 YHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHG 560
+H H +H + +HH + H H H + H H
Sbjct: 175 HHHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHGHHHHHHHHHGH 234
Query: 559 PHDGDNVHGHHDEDCVHGHYDAEDVHGHHDE 467
H + HGHH H H+DA H HH +
Sbjct: 235 HHHHHHHHGHHHHH--HHHHDAHHHHHHHHD 263
Score = 36.6 bits (83), Expect = 0.13
Identities = 22/92 (23%), Positives = 31/92 (33%), Gaps = 2/92 (2%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQS--NHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDC 569
++H H E H + +HH G + H+ + H H + H H
Sbjct: 136 HHHAAHHHHHEEHHHHHHAAHHHPWFHHHHLGYHHHHAPHHHHHHHHAPHHHHHHHHAPH 195
Query: 568 VHGPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
H H H HH H H+ H HH
Sbjct: 196 HHHHHHHAPHHHHHHHHAPHHHHHHHHGHHHH 227
Score = 35.0 bits (79), Expect = 0.39
Identities = 22/91 (24%), Positives = 39/91 (42%), Gaps = 2/91 (2%)
Frame = -3
Query: 739 YHIDFHDDFENHD-DQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDV-HGPHDEDCV 566
+H H++ +H ++ ++ + +HH + H + H PH + + H H
Sbjct: 69 HHEPHHEEHHHHHPEEHHEPHHEEHHHHHPHPHHHH---HHHPPHHHHHLGHHHHHHHAA 125
Query: 565 HGPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
H H + H HH H H+ E+ H HH
Sbjct: 126 HHHHHEE--HHHHHHAAHHHHH--EEHHHHH 152
Score = 35.0 bits (79), Expect = 0.39
Identities = 24/93 (25%), Positives = 33/93 (35%), Gaps = 3/93 (3%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGP--HDEDC 569
++H H + +H Y +HH H + H PH + H H
Sbjct: 151 HHHAAHHHPWFHHHHLGYHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHH 210
Query: 568 VHGPHDGDNVH-GHHDEDCVHGHYDAEDVHGHH 473
H PH + H GHH H H+ H HH
Sbjct: 211 HHAPHHHHHHHHGHHHHH--HHHHGHHHHHHHH 241
Score = 34.7 bits (78), Expect = 0.50
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 3/93 (3%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVH 563
++H + H + +H++ + +HH + H + + H H + H H E+ H
Sbjct: 79 HHHPEEHHE-PHHEEHHHHHPHPHHHHHHHPPHHHHHLGHHHHHH--HAAHHHHHEEHHH 135
Query: 562 GPHDGDNVHGHHDEDCVHGHYDAED---VHGHH 473
H H HH E+ H H+ A H HH
Sbjct: 136 HHHAA---HHHHHEEHHHHHHAAHHHPWFHHHH 165
Score = 34.3 bits (77), Expect = 0.66
Identities = 21/92 (22%), Positives = 31/92 (33%), Gaps = 3/92 (3%)
Frame = -3
Query: 739 YHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDP---HDVYDVHGPHDEDC 569
+H H +H +++ + HH + + H H P H H H
Sbjct: 117 HHHHHHHAAHHHHHEEHHHHHHAAHHHHHEEHHHHHHAAHHHPWFHHHHLGYHHHHAPHH 176
Query: 568 VHGPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
H H + H HH H H+ H HH
Sbjct: 177 HHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHH 208
Score = 33.1 bits (74), Expect = 1.5
Identities = 18/90 (20%), Positives = 32/90 (35%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVH 563
++H+ H +H + + +HH + H ++ H H + H +
Sbjct: 112 HHHLGHH---HHHHHAAHHHHHEEHHHHHHAAHHHHHEEHHHHHHAAHHHPWFHHHHLGY 168
Query: 562 GPHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
H + H HH H H+ H HH
Sbjct: 169 HHHHAPHHHHHHHHAPHHHHHHHHAPHHHH 198
>sp|Q9V3A4|CSUP_DROME Catecholamines up protein Length = 449 Score = 48.5 bits (114), Expect = 3e-05 Identities = 33/83 (39%), Positives = 39/83 (46%), Gaps = 1/83 (1%) Frame = -3 Query: 712 ENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGDNVHG 533 EN D QK + +HHD +DH D + H HD HD D HG G + HG Sbjct: 52 ENFDPQKAPRAEHHHHHD----HDH-DHGHHHHGHD-------HDHDHDHGHDHGHHHHG 99 Query: 532 H-HDEDCVHGHYDAEDVHGHHDE 467 H HD D HGH+ H HDE Sbjct: 100 HDHDHDHDHGHH-----HHGHDE 117
>sp|P05227|HRP1_PLAFA Histidine-rich protein precursor (Clone PFHRP-II) Length = 332 Score = 48.1 bits (113), Expect = 4e-05 Identities = 25/85 (29%), Positives = 31/85 (36%), Gaps = 1/85 (1%) Frame = -3 Query: 724 HDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDE-DCVHGPHDG 548 H D +H D + H D + + H H D H H D H H Sbjct: 53 HVDDAHHAHHVADAHHAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAA 112 Query: 547 DNVHGHHDEDCVHGHYDAEDVHGHH 473 D H HH D H H+ A+ H HH Sbjct: 113 DAHHAHHAADAHHAHHAADAHHAHH 137
Score = 47.4 bits (111), Expect = 8e-05
Identities = 24/73 (32%), Positives = 30/73 (41%), Gaps = 2/73 (2%)
Frame = -3
Query: 685 IQDQSNHHDDGDIYDH-SDVDNVHDPHDVYDV-HGPHDEDCVHGPHDGDNVHGHHDEDCV 512
+ + H DD H +D + H H D H H D H H D H HH D
Sbjct: 47 LHETQAHVDDAHHAHHVADAHHAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADAH 106
Query: 511 HGHYDAEDVHGHH 473
H H+ A+ H HH
Sbjct: 107 HAHHAADAHHAHH 119
Score = 47.4 bits (111), Expect = 8e-05
Identities = 25/89 (28%), Positives = 32/89 (35%), Gaps = 1/89 (1%)
Frame = -3
Query: 736 HIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDE-DCVHG 560
H H +H + D + H D + + H H D H H D H
Sbjct: 58 HHAHHVADAHHAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHA 117
Query: 559 PHDGDNVHGHHDEDCVHGHYDAEDVHGHH 473
H D H HH D H H+ A+ H HH
Sbjct: 118 HHAADAHHAHHAADAHHAHHAADAHHAHH 146
Score = 45.4 bits (106), Expect = 3e-04
Identities = 22/65 (33%), Positives = 25/65 (38%)
Frame = -3
Query: 667 HHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGDNVHGHHDEDCVHGHYDAED 488
HH + H D H H H H D H H D H HH D H H+ A+
Sbjct: 91 HHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADA 150
Query: 487 VHGHH 473
H HH
Sbjct: 151 HHAHH 155
Score = 43.9 bits (102), Expect = 8e-04
Identities = 22/65 (33%), Positives = 24/65 (36%)
Frame = -3
Query: 667 HHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGDNVHGHHDEDCVHGHYDAED 488
HH + H D H H H H D H H D H HH D H H+ A
Sbjct: 100 HHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAAYA 159
Query: 487 VHGHH 473
H HH
Sbjct: 160 HHAHH 164
Score = 37.4 bits (85), Expect = 0.078
Identities = 23/93 (24%), Positives = 31/93 (33%), Gaps = 3/93 (3%)
Frame = -3
Query: 742 YYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVH 563
Y H H +H + D + H D + +D + D H D H H D H
Sbjct: 200 YAHHAHHAADAHHAADAHHATDAHHAHHAADAHHATDAHHAADAHHAADAH--HATDAHH 257
Query: 562 GPHDGDNVHGHHDEDCVH---GHYDAEDVHGHH 473
HH D H H+ + H HH
Sbjct: 258 AADAHHATDAHHAADAHHAADAHHATDSHHAHH 290
Score = 35.8 bits (81), Expect = 0.23
Identities = 22/69 (31%), Positives = 24/69 (34%), Gaps = 4/69 (5%)
Frame = -3
Query: 667 HHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGDNVHGHHDEDCVHGHYDAED 488
HH + H D H H H H D H H D H HH H H+ A D
Sbjct: 109 HHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAAYAHHAHH-ASD 167
Query: 487 VH----GHH 473
H HH
Sbjct: 168 AHHAADAHH 176
Score = 35.8 bits (81), Expect = 0.23
Identities = 22/86 (25%), Positives = 29/86 (33%), Gaps = 2/86 (2%)
Frame = -3
Query: 724 HDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGD 545
H +H D ++ H D + +D + D H D H H D H
Sbjct: 218 HATDAHHAHHAADAHHATDAHHAADAHHAADAHHATDAHHAADAH--HATDAHHAADAHH 275
Query: 544 NVHGHHDEDCVHGHY--DAEDVHGHH 473
HH D H H+ DA HH
Sbjct: 276 AADAHHATDSHHAHHAADAHHAAAHH 301
Score = 35.4 bits (80), Expect = 0.30
Identities = 21/68 (30%), Positives = 23/68 (33%), Gaps = 3/68 (4%)
Frame = -3
Query: 667 HHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGDNVHGHHDEDCVH---GHYD 497
HH + H D H H H H D H H H HH D H H+
Sbjct: 118 HHAADAHHAHHAADAHHAHHAADAHHAHHAADAHHAHHAAYAHHAHHASDAHHAADAHHA 177
Query: 496 AEDVHGHH 473
A H HH
Sbjct: 178 AYAHHAHH 185
Score = 35.0 bits (79), Expect = 0.39
Identities = 24/101 (23%), Positives = 37/101 (36%), Gaps = 4/101 (3%)
Frame = -3
Query: 760 YNAIIFYYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPH 581
++A ++ D H + H D ++ H D + +D + D H D H
Sbjct: 226 HHAADAHHATDAHHAADAH--HAADAHHATDAHHAADAHHATDAHHAADAHHAADAHHAT 283
Query: 580 DEDCVHGPHDGDNVHGHHDEDCVH--GHY--DAEDVHGHHD 470
D H D + HH D H H+ DA HH+
Sbjct: 284 DSHHAHHAADAHHAAAHHATDAHHAAAHHATDAHHAAAHHE 324
>sp|P08723|SPBP_RAT Prostatic spermine-binding protein precursor (SBP) Length = 279 Score = 45.8 bits (107), Expect = 2e-04 Identities = 26/71 (36%), Positives = 36/71 (50%) Frame = -3 Query: 730 DFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHD 551 + HDDF+++DD K D D+ HDD + DH D DN +D D +D ED D Sbjct: 151 NIHDDFDDNDDDKEDDDDE---HDDDNEEDHGDKDNDNDHDDDHDDDDDDKED--DNEED 205 Query: 550 GDNVHGHHDED 518 D+ D+D Sbjct: 206 VDDERDDKDDD 216
Score = 30.4 bits (67), Expect = 9.5
Identities = 28/82 (34%), Positives = 33/82 (40%), Gaps = 1/82 (1%)
Frame = -3
Query: 730 DFHDDFENHDDQKYDIQDQSNHHDDGD-IYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPH 554
D DD E D + D +D DD D D D + D D D D+D G
Sbjct: 197 DKEDDNEEDVDDERDDKDDDEEDDDNDKENDKDDGEGSGDDDDNDDEDDDKDDDGGSG-D 255
Query: 553 DGDNVHGHHDEDCVHGHYDAED 488
DGD+ G DED G D D
Sbjct: 256 DGDD--GDDDEDDDGGDDDNGD 275
>sp|Q92504|KE4_HUMAN Zinc transporter SLC39A7 (Solute carrier family 39 member 7) (Histidine-rich membrane protein Ke4) Length = 469 Score = 45.4 bits (106), Expect = 3e-04 Identities = 33/93 (35%), Positives = 37/93 (39%), Gaps = 7/93 (7%) Frame = -3 Query: 724 HDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGD 545 HDD HDD + D S+ H D H H HG E HG H D Sbjct: 31 HDDL--HDDLQEDFHGHSHRHSHEDF---------HHGHSHAHGHGHTHESIWHG-HTHD 78 Query: 544 NVHGHHDEDCVHGH---YDAEDV----HGHHDE 467 + HGH ED HGH Y E + HGH E Sbjct: 79 HDHGHSHEDLHHGHSHGYSHESLYHRGHGHDHE 111
>sp|P04196|HRG_HUMAN Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG) Length = 525 Score = 42.4 bits (98), Expect = 0.002 Identities = 25/79 (31%), Positives = 37/79 (46%) Frame = -3 Query: 709 NHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGDNVHGH 530 +H++ D+ +H + + H + H PH+ +D H H H PH G + HGH Sbjct: 341 SHNNNSSDLHPHKHHSHEQ--HPHGHHPHAHHPHE-HDTHRQHPHG--HHPH-GHHPHGH 394 Query: 529 HDEDCVHGHYDAEDVHGHH 473 H HGH+ HGHH Sbjct: 395 HP----HGHHP----HGHH 405
Score = 41.6 bits (96), Expect = 0.004
Identities = 22/71 (30%), Positives = 33/71 (46%), Gaps = 1/71 (1%)
Frame = -3
Query: 682 QDQSNHHDDGDIYDHSDVDNVHDPHDVYD-VHGPHDEDCVHGPHDGDNVHGHHDEDCVHG 506
Q S++++ D++ H + PH + H PH+ D G + HGHH HG
Sbjct: 338 QRHSHNNNSSDLHPHKHHSHEQHPHGHHPHAHHPHEHDTHRQHPHGHHPHGHHP----HG 393
Query: 505 HYDAEDVHGHH 473
H+ HGHH
Sbjct: 394 HHP----HGHH 400
Score = 30.4 bits (67), Expect = 9.5
Identities = 22/85 (25%), Positives = 29/85 (34%), Gaps = 7/85 (8%)
Frame = -3
Query: 736 HIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGP 557
H H E H + + HD + H + H PH + HG H H P
Sbjct: 350 HPHKHHSHEQHPHGHHPHAHHPHEHDTHRQHPHGHHPHGHHPHG-HHPHGHHPHG--HHP 406
Query: 556 HDGD-------NVHGHHDEDCVHGH 503
H D + H+ C HGH
Sbjct: 407 HCHDFQDYGPCDPPPHNQGHCCHGH 431
>sp|Q31125|KE4_MOUSE Zinc transporter SLC39A7 (Solute carrier family 39 member 7) (Histidine-rich membrane protein Ke4) Length = 476 Score = 41.2 bits (95), Expect = 0.005 Identities = 27/82 (32%), Positives = 34/82 (41%) Frame = -3 Query: 712 ENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGDNVHG 533 E H D D+++ + H G + H D + H HG ED HG HG Sbjct: 29 EGHGDLHKDVEEDFHGHSHG--HSHEDFHHGHS-------HGHSHEDFHHG-------HG 72 Query: 532 HHDEDCVHGHYDAEDVHGHHDE 467 H E HGH + D HGH E Sbjct: 73 HTHESIWHGHAHSHD-HGHSRE 93
>sp|P08016|EGGS_SCHMA Putative eggshell protein Length = 149 Score = 40.0 bits (92), Expect = 0.012 Identities = 24/91 (26%), Positives = 36/91 (39%) Frame = -3 Query: 739 YHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHG 560 Y D H ++++ Y + N+ D YD D + ++ + H HD D Sbjct: 60 YGDDKHGHGKDYEKYGYTKEYSKNYKDYYKKYDKYDYGSRYEKYSYRKDHDKHDHD---- 115 Query: 559 PHDGDNVHGHHDEDCVHGHYDAEDVHGHHDE 467 H HHD+ H H+ E H HH E Sbjct: 116 ------EHDHHDDHHDHRHHHHEHDHHHHHE 140
Score = 33.9 bits (76), Expect = 0.86
Identities = 27/93 (29%), Positives = 36/93 (38%), Gaps = 13/93 (13%)
Frame = -3
Query: 742 YYHIDFHDDFENH--DDQKY-----------DIQDQSNHHDDGDIYDHSDVDNVHDPHDV 602
Y + + DD H D +KY D + + +D G Y+ HD HD
Sbjct: 55 YGYDKYGDDKHGHGKDYEKYGYTKEYSKNYKDYYKKYDKYDYGSRYEKYSYRKDHDKHD- 113
Query: 601 YDVHGPHDEDCVHGPHDGDNVHGHHDEDCVHGH 503
HDE H H D+ H HH+ D H H
Sbjct: 114 ------HDEHDHHDDHH-DHRHHHHEHDHHHHH 139
>sp|O67204|YB27_AQUAE Hypothetical protein AQ_1127 Length = 122 Score = 39.7 bits (91), Expect = 0.016 Identities = 28/90 (31%), Positives = 47/90 (52%), Gaps = 5/90 (5%) Frame = -3 Query: 769 SKIYNAIIFYYHIDFHDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVH 590 ++ ++ + Y H + +D N D++K D+ + H ++D D D+VHD D Y +H Sbjct: 12 TRTFHFLRSYGHDGWRNDGRN-DERKADV----SAHVLLPLHDAYDADDVHDADDGYALH 66 Query: 589 GPHDEDCVHGPHDGDNVH-----GHHDEDC 515 G + PHDG++ H G+H EDC Sbjct: 67 G-------YEPHDGNDEHADDGSGNH-EDC 88
Score = 32.3 bits (72), Expect = 2.5
Identities = 16/48 (33%), Positives = 25/48 (52%)
Frame = -3
Query: 610 HDVYDVHGPHDEDCVHGPHDGDNVHGHHDEDCVHGHYDAEDVHGHHDE 467
H + +H +D D VH DG +HG+ D H A+D G+H++
Sbjct: 42 HVLLPLHDAYDADDVHDADDGYALHGYEPHDGNDEH--ADDGSGNHED 87
>sp|Q9XTQ7|KE4L_CAEEL Hypothetical Ke4-like protein H13N06.5 in chromosome X Length = 515 Score = 38.1 bits (87), Expect = 0.046 Identities = 26/68 (38%), Positives = 32/68 (47%), Gaps = 2/68 (2%) Frame = -3 Query: 664 HDDGDIYDHSDVDNVHDPHDVYDVHG-PHDEDCVHGPHDGDNVHGH-HDEDCVHGHYDAE 491 HD G +DH D + HD +D HG HDE+ D+ HGH HD HGH + Sbjct: 129 HDHGHAHDH-DHGHAHDHGHAHDHHGHSHDEE-------EDHHHGHAHDH---HGHSHED 177 Query: 490 DVHGHHDE 467 H H E Sbjct: 178 HGHSHGAE 185
Score = 36.2 bits (82), Expect = 0.17
Identities = 27/90 (30%), Positives = 32/90 (35%), Gaps = 20/90 (22%)
Frame = -3
Query: 676 QSNHHDDGDIYDHSDVDNVHDPHD----------------VYDVHGPHDEDCVHGPHDGD 545
Q N H G + H + HD +D HD D H HD
Sbjct: 88 QQNVHHQGHGHAHGGHGHAHDADGGCPYAKAAAAEAATAAAHDHGHAHDHDHGHA-HDHG 146
Query: 544 NVHGHH----DEDCVHGHYDAEDVHGHHDE 467
+ H HH DE+ H H A D HGH E
Sbjct: 147 HAHDHHGHSHDEEEDHHHGHAHDHHGHSHE 176
Score = 33.5 bits (75), Expect = 1.1
Identities = 24/74 (32%), Positives = 29/74 (39%)
Frame = -3
Query: 724 HDDFENHDDQKYDIQDQSNHHDDGDIYDHSDVDNVHDPHDVYDVHGPHDEDCVHGPHDGD 545
HD HD HD G +DH + HD H H +E+ H H D
Sbjct: 129 HDHGHAHD------------HDHGHAHDHG---HAHDHHG----HSHDEEEDHHHGHAHD 169
Query: 544 NVHGHHDEDCVHGH 503
+ HGH ED H H
Sbjct: 170 H-HGHSHEDHGHSH 182
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 93,117,543
Number of Sequences: 369166
Number of extensions: 1784204
Number of successful extensions: 6990
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4876
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6328
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 12249817620
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail