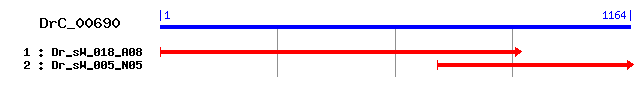
DrC_00690
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00690 (1164 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P15562|YMA6_PODAN Hypothetical 34.3 kDa protein in ATP6 ... 33 2.0 sp|Q04544|POLN_SOUV3 Non-structural polyprotein [Contains: ... 32 3.5 sp|Q12224|RLM1_YEAST Transcription factor RLM1 32 3.5 sp|Q6P0N0|CV106_HUMAN Protein C14orf106 (P243) 32 3.5 sp|Q9L7Q2|ZMPB_STRPN Zinc metalloprotease zmpB precursor 31 7.8
>sp|P15562|YMA6_PODAN Hypothetical 34.3 kDa protein in ATP6 5'region (ORFC) Length = 296 Score = 32.7 bits (73), Expect = 2.0 Identities = 21/84 (25%), Positives = 41/84 (48%), Gaps = 7/84 (8%) Frame = +2 Query: 479 LASLMVRLKEYDIRPKRVDSLRNSLFSL-----MFNDQSFSNDSDILKVFQNEPNLIHDP 643 L+S+ V + P ++ L N L + ++++QS I +V N+PN ++DP Sbjct: 193 LSSMSVEMPSNTTNPSDIERLNNLLEDIKKSLELYDNQSLKFRKHISEVNNNDPNCVYDP 252 Query: 644 LNIRRPRIYPNYLR--DYVIDNNK 709 R ++ +Y++ D + D K Sbjct: 253 ---RSKELFKDYIKLVDLLKDQQK 273
>sp|Q04544|POLN_SOUV3 Non-structural polyprotein [Contains: p48; Helicase (2C-like protein) (P2C); 3A-like protein; Viral genome-linked protein (VPg); Thiol protease P3C (3C-like protease) (3C-pro); RNA-directed RNA polymerase ] Length = 1788 Score = 32.0 bits (71), Expect = 3.5 Identities = 38/146 (26%), Positives = 62/146 (42%), Gaps = 22/146 (15%) Frame = +2 Query: 224 LTSVQELIERLARNKIIPNRMDCGI------DSTFTLFICDEQFSKISQYTNENLLIIRF 385 LT+ EL +A++ + P+ MD G + + F C Q + I+ + Sbjct: 1544 LTASPELASVVAQDLLAPSEMDVGDYVIRVKEGLPSGFPCTSQVNSINHW---------- 1593 Query: 386 LPHMAVVTECTNLSLRIENS----------EIQICSISYDSLASLMVRLKEYDIRPKRVD 535 L + ++E T LS + S EI I +D A L L+EY +RP R D Sbjct: 1594 LITLCALSEVTGLSPDVIQSMSYFSFYGDDEIVSTDIEFDP-AKLTQVLREYGLRPTRPD 1652 Query: 536 S------LRNSLFSLMFNDQSFSNDS 595 +R S+ L+F ++ S D+ Sbjct: 1653 KSEGPIIVRKSVDGLVFLRRTISRDA 1678
>sp|Q12224|RLM1_YEAST Transcription factor RLM1 Length = 676 Score = 32.0 bits (71), Expect = 3.5 Identities = 22/87 (25%), Positives = 45/87 (51%) Frame = +2 Query: 566 FNDQSFSNDSDILKVFQNEPNLIHDPLNIRRPRIYPNYLRDYVIDNNKGEPVSGLDLISF 745 F + S + +D++ +QN+ NL+H+ ++ P Y ++ + ++ N+ S + S Sbjct: 54 FYEFSSVDTNDLIYHYQNDKNLLHE---VKDPSDYGDFHKSASVNINQDLLRSSM---SN 107 Query: 746 VPSRSNFAPVKTSQNHSNNESLEDLGD 826 PS+SN + S+N + + ED D Sbjct: 108 KPSKSNVKGMNQSENDDDENNDEDDDD 134
>sp|Q6P0N0|CV106_HUMAN Protein C14orf106 (P243) Length = 1132 Score = 32.0 bits (71), Expect = 3.5 Identities = 22/88 (25%), Positives = 42/88 (47%) Frame = +2 Query: 332 QFSKISQYTNENLLIIRFLPHMAVVTECTNLSLRIENSEIQICSISYDSLASLMVRLKEY 511 QF K++ + N+N+ L ++S N + +Y+S + +R+KE Sbjct: 62 QFLKMTTFNNKNIFQSTMLTEATTSNSSLDISAIKPNKDGLKNKANYESPGKIFLRMKEK 121 Query: 512 DIRPKRVDSLRNSLFSLMFNDQSFSNDS 595 +R K+ RNS SL+ +S +N++ Sbjct: 122 VLRDKQEQPSRNS--SLLEPQKSGNNET 147
>sp|Q9L7Q2|ZMPB_STRPN Zinc metalloprotease zmpB precursor Length = 1906 Score = 30.8 bits (68), Expect = 7.8 Identities = 41/160 (25%), Positives = 75/160 (46%) Frame = +2 Query: 263 NKIIPNRMDCGIDSTFTLFICDEQFSKISQYTNENLLIIRFLPHMAVVTECTNLSLRIEN 442 NKII + D G F L E S + +YT +L I ++ P++ V + T L I+ Sbjct: 1142 NKIIVHYAD-GTKDYFNLSSSSEGLSNVKEYTITDLGI-KYTPNI-VQKDNTTLVNDIK- 1197 Query: 443 SEIQICSISYDSLASLMVRLKEYDIRPKRVDSLRNSLFSLMFNDQSFSNDSDILKVFQNE 622 S ++ + ++ + RL +Y RV+++++ F D + + I K+ QNE Sbjct: 1198 SILESVELQSQTMYQHLNRLGDY-----RVNAIKDLYLEESFTDVKENLTNLITKLVQNE 1252 Query: 623 PNLIHDPLNIRRPRIYPNYLRDYVIDNNKGEPVSGLDLIS 742 + ++D R+ +RD V + NK + GL ++ Sbjct: 1253 EHQLNDSPAARQ------MIRDKV-EKNKAALLLGLTYLN 1285
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 124,017,529
Number of Sequences: 369166
Number of extensions: 2418795
Number of successful extensions: 5580
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 5385
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5578
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 13107781500
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail