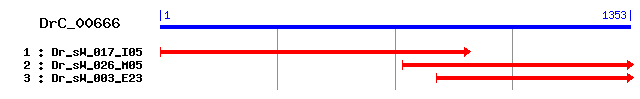
DrC_00666
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00666 (1353 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P61980|HNRPK_RAT Heterogeneous nuclear ribonucleoprotein... 89 4e-17 sp|O19049|HNRPK_RABIT Heterogeneous nuclear ribonucleoprote... 89 4e-17 sp|P57722|PCBP3_MOUSE Poly(rC)-binding protein 3 (Alpha-CP3) 57 2e-07 sp|P57721|PCBP3_HUMAN Poly(rC)-binding protein 3 (Alpha-CP3) 57 2e-07 sp|P57723|PCBP4_HUMAN Poly(rC)-binding protein 4 (Alpha-CP4) 55 4e-07 sp|P60335|PCBP1_MOUSE Poly(rC)-binding protein 1 (Alpha-CP1... 52 4e-06 sp|P57724|PCBP4_MOUSE Poly(rC)-binding protein 4 (Alpha-CP4) 52 5e-06 sp|Q15366|PCBP2_HUMAN Poly(rC)-binding protein 2 (Alpha-CP2... 51 7e-06 sp|Q61990|PCBP2_MOUSE Poly(rC)-binding protein 2 (Alpha-CP2... 51 7e-06 sp|Q9UNW9|NOVA2_HUMAN RNA-binding protein Nova-2 (Neuro-onc... 49 3e-05
>sp|P61980|HNRPK_RAT Heterogeneous nuclear ribonucleoprotein K (dC stretch-binding protein) (CSBP) sp|P61979|HNRPK_MOUSE Heterogeneous nuclear ribonucleoprotein K sp|P61978|HNRPK_HUMAN Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP) Length = 463 Score = 88.6 bits (218), Expect = 4e-17 Identities = 45/87 (51%), Positives = 61/87 (70%) Frame = +3 Query: 1041 SMGSHSGPIEISHVSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIIT 1220 S G GPI + V++P L G+I+GKGG RI +IR E+GA I +D+ + S +RIIT Sbjct: 379 SYGDLGGPIITTQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPL--EGSEDRIIT 436 Query: 1221 IQGSRDQIQNAQFLLQACVRKYGGKHF 1301 I G++DQIQNAQ+LLQ V++Y GK F Sbjct: 437 ITGTQDQIQNAQYLLQNSVKQYSGKFF 463
Score = 56.6 bits (135), Expect = 2e-07
Identities = 26/62 (41%), Positives = 41/62 (66%)
Frame = +1
Query: 40 LQYRFLIPQPAAGAIISKKGETIKRLRATYNCDISVPPSNGPERLMIINTEIDRLCGVLG 219
++ R L+ AGA+I K G+ IK LR YN +SVP S+GPER++ I+ +I+ + +L
Sbjct: 43 VELRILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSGPERILSISADIETIGEILK 102
Query: 220 EI 225
+I
Sbjct: 103 KI 104
Score = 41.2 bits (95), Expect = 0.007
Identities = 15/39 (38%), Positives = 29/39 (74%)
Frame = +2
Query: 536 SVIKVYQQMCPSSTDRVIRLVGLPEDILSCLRAIHEVIT 652
+ IK++Q+ CP STDRV+ + G P+ ++ C++ I ++I+
Sbjct: 176 TTIKLFQECCPHSTDRVVLIGGKPDRVVECIKIILDLIS 214
Score = 35.0 bits (79), Expect = 0.50
Identities = 24/88 (27%), Positives = 38/88 (43%)
Frame = +3
Query: 981 DLSQEQVYSNPTPTPLMPHLSMGSHSGPIEISHVSLPNKLIGAIMGKGGSRINEIREETG 1160
D+ +EQ + T M L + L +K GA++GKGG I +R +
Sbjct: 26 DMEEEQAFKRSRNTDEMVEL------------RILLQSKNAGAVIGKGGKNIKALRTDYN 73
Query: 1161 ARIDVDKTMMDNKSSERIITIQGSRDQI 1244
A + V D+ ERI++I + I
Sbjct: 74 ASVSVP----DSSGPERILSISADIETI 97
>sp|O19049|HNRPK_RABIT Heterogeneous nuclear ribonucleoprotein K (hnRNP K) Length = 463 Score = 88.6 bits (218), Expect = 4e-17 Identities = 45/87 (51%), Positives = 61/87 (70%) Frame = +3 Query: 1041 SMGSHSGPIEISHVSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIIT 1220 S G GPI + V++P L G+I+GKGG RI +IR E+GA I +D+ + S +RIIT Sbjct: 379 SYGDLGGPIITTQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPL--EGSEDRIIT 436 Query: 1221 IQGSRDQIQNAQFLLQACVRKYGGKHF 1301 I G++DQIQNAQ+LLQ V++Y GK F Sbjct: 437 ITGTQDQIQNAQYLLQNSVKQYSGKFF 463
Score = 56.6 bits (135), Expect = 2e-07
Identities = 26/62 (41%), Positives = 41/62 (66%)
Frame = +1
Query: 40 LQYRFLIPQPAAGAIISKKGETIKRLRATYNCDISVPPSNGPERLMIINTEIDRLCGVLG 219
++ R L+ AGA+I K G+ IK LR YN +SVP S+GPER++ I+ +I+ + +L
Sbjct: 43 VELRILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSGPERILSISADIETIGEILK 102
Query: 220 EI 225
+I
Sbjct: 103 KI 104
Score = 41.2 bits (95), Expect = 0.007
Identities = 15/39 (38%), Positives = 29/39 (74%)
Frame = +2
Query: 536 SVIKVYQQMCPSSTDRVIRLVGLPEDILSCLRAIHEVIT 652
+ IK++Q+ CP STDRV+ + G P+ ++ C++ I ++I+
Sbjct: 176 TTIKLFQECCPHSTDRVVLIGGKPDRVVECIKIILDLIS 214
Score = 35.0 bits (79), Expect = 0.50
Identities = 24/88 (27%), Positives = 38/88 (43%)
Frame = +3
Query: 981 DLSQEQVYSNPTPTPLMPHLSMGSHSGPIEISHVSLPNKLIGAIMGKGGSRINEIREETG 1160
D+ +EQ + T M L + L +K GA++GKGG I +R +
Sbjct: 26 DMEEEQAFKRSRNTDEMVEL------------RILLQSKNAGAVIGKGGKNIKALRTDYN 73
Query: 1161 ARIDVDKTMMDNKSSERIITIQGSRDQI 1244
A + V D+ ERI++I + I
Sbjct: 74 ASVSVP----DSSGPERILSISADIETI 97
>sp|P57722|PCBP3_MOUSE Poly(rC)-binding protein 3 (Alpha-CP3) Length = 339 Score = 56.6 bits (135), Expect = 2e-07 Identities = 34/81 (41%), Positives = 48/81 (59%), Gaps = 5/81 (6%) Frame = +3 Query: 1044 MGSHSG-----PIEISHVSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSE 1208 MG SG P +++PN LIG I+G+ G++INEIR+ +GA+I + SSE Sbjct: 249 MGQSSGLDASPPASTHELTIPNDLIGCIIGRQGTKINEIRQMSGAQIKIANA--TEGSSE 306 Query: 1209 RIITIQGSRDQIQNAQFLLQA 1271 R ITI G+ I AQ+L+ A Sbjct: 307 RQITITGTPANISLAQYLINA 327
Score = 53.9 bits (128), Expect = 1e-06
Identities = 30/64 (46%), Positives = 42/64 (65%)
Frame = +3
Query: 1086 LPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNAQFLL 1265
+P G+++GKGGS+I EIRE TGA++ V M+ N S+ER +TI G+ D I Q +
Sbjct: 104 VPASQCGSLIGKGGSKIKEIRESTGAQVQVAGDMLPN-STERAVTISGTPDAI--IQCVK 160
Query: 1266 QACV 1277
Q CV
Sbjct: 161 QICV 164
Score = 42.7 bits (99), Expect = 0.002
Identities = 22/53 (41%), Positives = 33/53 (62%)
Frame = +3
Query: 1095 KLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNA 1253
K +G+I+GK G + ++REE+GARI++ + ERI+TI G D I A
Sbjct: 23 KEVGSIIGKKGETVKKMREESGARINIS----EGNCPERIVTITGPTDAIFKA 71
Score = 40.8 bits (94), Expect = 0.009
Identities = 20/56 (35%), Positives = 31/56 (55%)
Frame = +1
Query: 37 SLQYRFLIPQPAAGAIISKKGETIKRLRATYNCDISVPPSNGPERLMIINTEIDRL 204
+L R L+ G+II KKGET+K++R I++ N PER++ I D +
Sbjct: 13 TLTIRLLMHGKEVGSIIGKKGETVKKMREESGARINISEGNCPERIVTITGPTDAI 68
Score = 32.7 bits (73), Expect = 2.5
Identities = 17/45 (37%), Positives = 27/45 (60%), Gaps = 1/45 (2%)
Frame = +3
Query: 345 KDIINGIMPDGIVSAD-MDLRFLVSENVCGAIIGKGGEVIRKLTE 476
+DIIN + S + LR +V + CG++IGKGG I+++ E
Sbjct: 81 EDIINSMSNSPATSKPPVTLRLVVPASQCGSLIGKGGSKIKEIRE 125
>sp|P57721|PCBP3_HUMAN Poly(rC)-binding protein 3 (Alpha-CP3) Length = 339 Score = 56.6 bits (135), Expect = 2e-07 Identities = 34/81 (41%), Positives = 48/81 (59%), Gaps = 5/81 (6%) Frame = +3 Query: 1044 MGSHSG-----PIEISHVSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSE 1208 MG SG P +++PN LIG I+G+ G++INEIR+ +GA+I + SSE Sbjct: 249 MGQSSGLDASPPASTHELTIPNDLIGCIIGRQGTKINEIRQMSGAQIKIANA--TEGSSE 306 Query: 1209 RIITIQGSRDQIQNAQFLLQA 1271 R ITI G+ I AQ+L+ A Sbjct: 307 RQITITGTPANISLAQYLINA 327
Score = 53.9 bits (128), Expect = 1e-06
Identities = 30/64 (46%), Positives = 42/64 (65%)
Frame = +3
Query: 1086 LPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNAQFLL 1265
+P G+++GKGGS+I EIRE TGA++ V M+ N S+ER +TI G+ D I Q +
Sbjct: 104 VPASQCGSLIGKGGSKIKEIRESTGAQVQVAGDMLPN-STERAVTISGTPDAI--IQCVK 160
Query: 1266 QACV 1277
Q CV
Sbjct: 161 QICV 164
Score = 42.7 bits (99), Expect = 0.002
Identities = 22/53 (41%), Positives = 33/53 (62%)
Frame = +3
Query: 1095 KLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNA 1253
K +G+I+GK G + ++REE+GARI++ + ERI+TI G D I A
Sbjct: 23 KEVGSIIGKKGETVKKMREESGARINIS----EGNCPERIVTITGPTDAIFKA 71
Score = 40.8 bits (94), Expect = 0.009
Identities = 20/56 (35%), Positives = 31/56 (55%)
Frame = +1
Query: 37 SLQYRFLIPQPAAGAIISKKGETIKRLRATYNCDISVPPSNGPERLMIINTEIDRL 204
+L R L+ G+II KKGET+K++R I++ N PER++ I D +
Sbjct: 13 TLTIRLLMHGKEVGSIIGKKGETVKKMREESGARINISEGNCPERIVTITGPTDAI 68
Score = 32.7 bits (73), Expect = 2.5
Identities = 17/45 (37%), Positives = 27/45 (60%), Gaps = 1/45 (2%)
Frame = +3
Query: 345 KDIINGIMPDGIVSAD-MDLRFLVSENVCGAIIGKGGEVIRKLTE 476
+DIIN + S + LR +V + CG++IGKGG I+++ E
Sbjct: 81 EDIINSMSNSPATSKPPVTLRLVVPASQCGSLIGKGGSKIKEIRE 125
>sp|P57723|PCBP4_HUMAN Poly(rC)-binding protein 4 (Alpha-CP4) Length = 403 Score = 55.5 bits (132), Expect = 4e-07 Identities = 36/99 (36%), Positives = 56/99 (56%), Gaps = 1/99 (1%) Frame = +3 Query: 984 LSQEQVYSNPTPTP-LMPHLSMGSHSGPIEISHVSLPNKLIGAIMGKGGSRINEIREETG 1160 L Q ++ P TP ++P L G+ + E +PN LIG ++G+ GS+I+EIR+ +G Sbjct: 216 LQQLSSHAVPFATPSVVPGLDPGTQTSSQEFL---VPNDLIGCVIGRQGSKISEIRQMSG 272 Query: 1161 ARIDVDKTMMDNKSSERIITIQGSRDQIQNAQFLLQACV 1277 A I + + ER +TI GS I AQ+L+ AC+ Sbjct: 273 AHIKIGN--QAEGAGERHVTITGSPVSIALAQYLITACL 309
Score = 47.4 bits (111), Expect = 1e-04
Identities = 25/66 (37%), Positives = 40/66 (60%)
Frame = +3
Query: 1047 GSHSGPIEISHVSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQ 1226
G+ S P + +P G+++GK G++I EIRE TGA++ V ++ N S+ER +T+
Sbjct: 95 GNVSRPPVTLRLVIPASQCGSLIGKAGTKIKEIRETTGAQVQVAGDLLPN-STERAVTVS 153
Query: 1227 GSRDQI 1244
G D I
Sbjct: 154 GVPDAI 159
Score = 38.5 bits (88), Expect = 0.045
Identities = 19/49 (38%), Positives = 29/49 (59%)
Frame = +1
Query: 37 SLQYRFLIPQPAAGAIISKKGETIKRLRATYNCDISVPPSNGPERLMII 183
+L R L+ G+II KKGET+KR+R + I++ + PER+ I
Sbjct: 17 TLTLRMLMHGKEVGSIIGKKGETVKRIREQSSARITISEGSCPERITTI 65
Score = 37.7 bits (86), Expect = 0.077
Identities = 21/53 (39%), Positives = 31/53 (58%)
Frame = +3
Query: 1095 KLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNA 1253
K +G+I+GK G + IRE++ ARI T+ + ERI TI GS + +A
Sbjct: 27 KEVGSIIGKKGETVKRIREQSSARI----TISEGSCPERITTITGSTAAVFHA 75
Score = 31.6 bits (70), Expect = 5.5
Identities = 13/36 (36%), Positives = 24/36 (66%)
Frame = +2
Query: 542 IKVYQQMCPSSTDRVIRLVGLPEDILSCLRAIHEVI 649
++V + P+ST+R + + G+P+ I+ C+R I VI
Sbjct: 135 VQVAGDLLPNSTERAVTVSGVPDAIILCVRQICAVI 170
>sp|P60335|PCBP1_MOUSE Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) sp|Q15365|PCBP1_HUMAN Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid-binding protein SUB2.3) sp|O19048|PCBP1_RABIT Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) Length = 356 Score = 52.0 bits (123), Expect = 4e-06 Identities = 28/64 (43%), Positives = 42/64 (65%) Frame = +3 Query: 1080 VSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNAQF 1259 +++PN LIG I+G+ G+ INEIR+ +GA+I + + SS R +TI GS I AQ+ Sbjct: 284 LTIPNNLIGCIIGRQGANINEIRQMSGAQIKIANPV--EGSSGRQVTITGSAASISLAQY 341 Query: 1260 LLQA 1271 L+ A Sbjct: 342 LINA 345
Score = 47.8 bits (112), Expect = 7e-05
Identities = 32/89 (35%), Positives = 51/89 (57%)
Frame = +3
Query: 963 ADSIMLDLSQEQVYSNPTPTPLMPHLSMGSHSGPIEISHVSLPNKLIGAIMGKGGSRINE 1142
A ++++D +E + S+ T S + P+ + V +P G+++GKGG +I E
Sbjct: 71 AFAMIIDKLEEDINSSMTN-------STAASRPPVTLRLV-VPATQCGSLIGKGGCKIKE 122
Query: 1143 IREETGARIDVDKTMMDNKSSERIITIQG 1229
IRE TGA++ V M+ N S+ER ITI G
Sbjct: 123 IRESTGAQVQVAGDMLPN-STERAITIAG 150
Score = 41.2 bits (95), Expect = 0.007
Identities = 22/53 (41%), Positives = 32/53 (60%)
Frame = +3
Query: 1095 KLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNA 1253
K +G+I+GK G + IREE+GARI++ + ERIIT+ G + I A
Sbjct: 23 KEVGSIIGKKGESVKRIREESGARINIS----EGNCPERIITLTGPTNAIFKA 71
Score = 38.5 bits (88), Expect = 0.045
Identities = 18/49 (36%), Positives = 29/49 (59%)
Frame = +1
Query: 37 SLQYRFLIPQPAAGAIISKKGETIKRLRATYNCDISVPPSNGPERLMII 183
+L R L+ G+II KKGE++KR+R I++ N PER++ +
Sbjct: 13 TLTIRLLMHGKEVGSIIGKKGESVKRIREESGARINISEGNCPERIITL 61
Score = 31.6 bits (70), Expect = 5.5
Identities = 11/32 (34%), Positives = 21/32 (65%)
Frame = +2
Query: 542 IKVYQQMCPSSTDRVIRLVGLPEDILSCLRAI 637
++V M P+ST+R I + G+P+ + C++ I
Sbjct: 131 VQVAGDMLPNSTERAITIAGVPQSVTECVKQI 162
>sp|P57724|PCBP4_MOUSE Poly(rC)-binding protein 4 (Alpha-CP4) Length = 403 Score = 51.6 bits (122), Expect = 5e-06 Identities = 27/64 (42%), Positives = 41/64 (64%) Frame = +3 Query: 1086 LPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNAQFLL 1265 +PN LIG ++G+ GS+I+EIR+ +GA I + + ER +TI GS I AQ+L+ Sbjct: 248 VPNDLIGCVIGRQGSKISEIRQMSGAHIKIGN--QAEGAGERHVTITGSPVSIALAQYLI 305 Query: 1266 QACV 1277 AC+ Sbjct: 306 TACL 309
Score = 48.5 bits (114), Expect = 4e-05
Identities = 26/66 (39%), Positives = 40/66 (60%)
Frame = +3
Query: 1047 GSHSGPIEISHVSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQ 1226
GS S P + +P G+++GK G++I EIRE TGA++ V ++ N S+ER +T+
Sbjct: 95 GSVSRPPVTLRLVIPASQCGSLIGKAGTKIKEIRETTGAQVQVAGDLLPN-STERAVTVS 153
Query: 1227 GSRDQI 1244
G D I
Sbjct: 154 GVPDAI 159
Score = 38.5 bits (88), Expect = 0.045
Identities = 19/49 (38%), Positives = 29/49 (59%)
Frame = +1
Query: 37 SLQYRFLIPQPAAGAIISKKGETIKRLRATYNCDISVPPSNGPERLMII 183
+L R L+ G+II KKGET+KR+R + I++ + PER+ I
Sbjct: 17 TLTLRMLMHGKEVGSIIGKKGETVKRIREQSSARITISEGSCPERITTI 65
Score = 37.7 bits (86), Expect = 0.077
Identities = 21/53 (39%), Positives = 31/53 (58%)
Frame = +3
Query: 1095 KLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNA 1253
K +G+I+GK G + IRE++ ARI T+ + ERI TI GS + +A
Sbjct: 27 KEVGSIIGKKGETVKRIREQSSARI----TISEGSCPERITTITGSTAAVFHA 75
Score = 31.6 bits (70), Expect = 5.5
Identities = 13/36 (36%), Positives = 24/36 (66%)
Frame = +2
Query: 542 IKVYQQMCPSSTDRVIRLVGLPEDILSCLRAIHEVI 649
++V + P+ST+R + + G+P+ I+ C+R I VI
Sbjct: 135 VQVAGDLLPNSTERAVTVSGVPDAIILCVRQICAVI 170
>sp|Q15366|PCBP2_HUMAN Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2) Length = 365 Score = 51.2 bits (121), Expect = 7e-06 Identities = 26/62 (41%), Positives = 43/62 (69%) Frame = +3 Query: 1080 VSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNAQF 1259 +++PN LIG I+G+ G++INEIR+ +GA+I + + S++R +TI GS I AQ+ Sbjct: 292 LTIPNDLIGCIIGRQGAKINEIRQMSGAQIKIANPV--EGSTDRQVTITGSAASISLAQY 349 Query: 1260 LL 1265 L+ Sbjct: 350 LI 351
Score = 48.9 bits (115), Expect = 3e-05
Identities = 36/105 (34%), Positives = 57/105 (54%)
Frame = +3
Query: 963 ADSIMLDLSQEQVYSNPTPTPLMPHLSMGSHSGPIEISHVSLPNKLIGAIMGKGGSRINE 1142
A ++++D +E + S+ T S + P+ + V +P G+++GKGG +I E
Sbjct: 71 AFAMIIDKLEEDISSSMTN-------STAASRPPVTLRLV-VPASQCGSLIGKGGCKIKE 122
Query: 1143 IREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNAQFLLQACV 1277
IRE TGA++ V M+ N S+ER ITI G I + + Q CV
Sbjct: 123 IRESTGAQVQVAGDMLPN-STERAITIAGIPQSI--IECVKQICV 164
Score = 40.4 bits (93), Expect = 0.012
Identities = 21/53 (39%), Positives = 33/53 (62%)
Frame = +3
Query: 1095 KLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNA 1253
K +G+I+GK G + ++REE+GARI++ + ERIIT+ G + I A
Sbjct: 23 KEVGSIIGKKGESVKKMREESGARINIS----EGNCPERIITLAGPTNAIFKA 71
Score = 37.4 bits (85), Expect = 0.10
Identities = 17/49 (34%), Positives = 29/49 (59%)
Frame = +1
Query: 37 SLQYRFLIPQPAAGAIISKKGETIKRLRATYNCDISVPPSNGPERLMII 183
+L R L+ G+II KKGE++K++R I++ N PER++ +
Sbjct: 13 TLTIRLLMHGKEVGSIIGKKGESVKKMREESGARINISEGNCPERIITL 61
Score = 33.5 bits (75), Expect = 1.5
Identities = 12/32 (37%), Positives = 22/32 (68%)
Frame = +2
Query: 542 IKVYQQMCPSSTDRVIRLVGLPEDILSCLRAI 637
++V M P+ST+R I + G+P+ I+ C++ I
Sbjct: 131 VQVAGDMLPNSTERAITIAGIPQSIIECVKQI 162
>sp|Q61990|PCBP2_MOUSE Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP) Length = 362 Score = 51.2 bits (121), Expect = 7e-06 Identities = 26/62 (41%), Positives = 43/62 (69%) Frame = +3 Query: 1080 VSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNAQF 1259 +++PN LIG I+G+ G++INEIR+ +GA+I + + S++R +TI GS I AQ+ Sbjct: 289 LTIPNDLIGCIIGRQGAKINEIRQMSGAQIKIANPV--EGSTDRQVTITGSAASISLAQY 346 Query: 1260 LL 1265 L+ Sbjct: 347 LI 348
Score = 48.9 bits (115), Expect = 3e-05
Identities = 36/105 (34%), Positives = 57/105 (54%)
Frame = +3
Query: 963 ADSIMLDLSQEQVYSNPTPTPLMPHLSMGSHSGPIEISHVSLPNKLIGAIMGKGGSRINE 1142
A ++++D +E + S+ T S + P+ + V +P G+++GKGG +I E
Sbjct: 71 AFAMIIDKLEEDISSSMTN-------STAASRPPVTLRLV-VPASQCGSLIGKGGCKIKE 122
Query: 1143 IREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNAQFLLQACV 1277
IRE TGA++ V M+ N S+ER ITI G I + + Q CV
Sbjct: 123 IRESTGAQVQVAGDMLPN-STERAITIAGIPQSI--IECVKQICV 164
Score = 40.4 bits (93), Expect = 0.012
Identities = 21/53 (39%), Positives = 33/53 (62%)
Frame = +3
Query: 1095 KLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNA 1253
K +G+I+GK G + ++REE+GARI++ + ERIIT+ G + I A
Sbjct: 23 KEVGSIIGKKGESVKKMREESGARINIS----EGNCPERIITLAGPTNAIFKA 71
Score = 37.4 bits (85), Expect = 0.10
Identities = 17/49 (34%), Positives = 29/49 (59%)
Frame = +1
Query: 37 SLQYRFLIPQPAAGAIISKKGETIKRLRATYNCDISVPPSNGPERLMII 183
+L R L+ G+II KKGE++K++R I++ N PER++ +
Sbjct: 13 TLTIRLLMHGKEVGSIIGKKGESVKKMREESGARINISEGNCPERIITL 61
Score = 33.5 bits (75), Expect = 1.5
Identities = 12/32 (37%), Positives = 22/32 (68%)
Frame = +2
Query: 542 IKVYQQMCPSSTDRVIRLVGLPEDILSCLRAI 637
++V M P+ST+R I + G+P+ I+ C++ I
Sbjct: 131 VQVAGDMLPNSTERAITIAGIPQSIIECVKQI 162
>sp|Q9UNW9|NOVA2_HUMAN RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein) Length = 492 Score = 48.9 bits (115), Expect = 3e-05 Identities = 27/67 (40%), Positives = 40/67 (59%), Gaps = 1/67 (1%) Frame = +3 Query: 1068 EISHVSLPNKLIGAIMGKGGSRINEIREETGARIDVDKT-MMDNKSSERIITIQGSRDQI 1244 E+ +++P L+GAI+GKGG + E +E TGARI + K + R +TI GS Sbjct: 407 ELVEIAVPENLVGAILGKGGKTLVEYQELTGARIQISKKGEFLPGTRNRRVTITGSPAAT 466 Query: 1245 QNAQFLL 1265 Q AQ+L+ Sbjct: 467 QAAQYLI 473
Score = 43.1 bits (100), Expect = 0.002
Identities = 17/56 (30%), Positives = 32/56 (57%)
Frame = +3
Query: 1086 LPNKLIGAIMGKGGSRINEIREETGARIDVDKTMMDNKSSERIITIQGSRDQIQNA 1253
+PN G I+GKGG+ + + E++GA + + + ER++T+ G +Q+ A
Sbjct: 137 VPNSTAGLIIGKGGATVKAVMEQSGAWVQLSQKPEGINLQERVVTVSGEPEQVHKA 192
Score = 39.7 bits (91), Expect = 0.020
Identities = 21/69 (30%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Frame = +3
Query: 1080 VSLPNKLIGAIMGKGGSRINEIREETGARIDVDKTM-MDNKSSERIITIQGSRDQIQNAQ 1256
V +P+ G+I+GKGG I ++++ETGA I + K+ ++ER+ +QG+ + +
Sbjct: 37 VLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKDFYPGTTERVCLVQGTAEALNAVH 96
Query: 1257 FLLQACVRK 1283
+ VR+
Sbjct: 97 SFIAEKVRE 105
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 148,180,946
Number of Sequences: 369166
Number of extensions: 3003976
Number of successful extensions: 7358
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6838
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7348
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 15891183840
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail