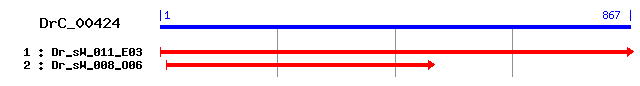
DrC_00424
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00424 (867 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P14272|KLKB1_RAT Plasma kallikrein precursor (Plasma pre... 102 1e-21 sp|O15393|TMPS2_HUMAN Transmembrane protease, serine 2 prec... 99 2e-20 sp|P26262|KLKB1_MOUSE Plasma kallikrein precursor (Plasma p... 97 8e-20 sp|Q9JIQ8|TMPS2_MOUSE Transmembrane protease, serine 2 (Epi... 91 4e-18 sp|Q7Z410|TMPS9_HUMAN Transmembrane protease, serine 9 (Pol... 87 5e-17 sp|P03952|KLKB1_HUMAN Plasma kallikrein precursor (Plasma p... 86 1e-16 sp|P98074|ENTK_PIG Enteropeptidase precursor (Enterokinase)... 83 9e-16 sp|P56677|ST14_MOUSE Suppressor of tumorigenicity 14 (Epithin) 83 1e-15 sp|Q9ER04|TMPS5_MOUSE Transmembrane protease, serine 5 (Spi... 82 2e-15 sp|P23578|ACRO_MOUSE Acrosin precursor [Contains: Acrosin l... 82 3e-15
>sp|P14272|KLKB1_RAT Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain] Length = 638 Score = 102 bits (255), Expect = 1e-21 Identities = 81/294 (27%), Positives = 126/294 (42%), Gaps = 5/294 (1%) Frame = +1 Query: 1 FFLCLSTITLAFKTNAKIMKRVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTK 180 + L L + + KI R++GG + ++ W VS++ V + + CG SI+ + Sbjct: 370 YSLRLCKVVESSDCTTKINARIVGGTNSSLGEWPWQVSLQ--VKLVSQNHMCGGSIIGRQ 427 Query: 181 GNISWLVTAAHCFYPSNKKKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFF 360 W++TAAHCF + P++W G++ + I NK T F Sbjct: 428 ----WILTAAHCFD--------GIPYPDVWRIYGGILNLSEITNK------------TPF 463 Query: 361 SKAFRKLFQSKQNEVVTTGDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNT 540 S IK +IIH YK + YDIAL+KL PL + + Sbjct: 464 SS--------------------IKELIIHQKYKMSEGSYDIALIKLQTPLNYTEFQKPIC 503 Query: 541 IDMPDIPKGVSWPEVDSECVVVGWGCTKAAGVPNNIAKLAKLKVWDNQKCIDAFVAPIDL 720 + P + + C V GWG TK G NI + A + + N++C + D Sbjct: 504 L-----PSKADTNTIYTNCWVTGWGYTKERGETQNILQKATIPLVPNEECQKKYR---DY 555 Query: 721 NPTSEF-CAGYLKKKYGLCPGDSGSGLACQYNGKWTFAGVVS----AAHSKEPG 867 T + CAGY + C GDSG L C+++G+W G+ S A ++PG Sbjct: 556 VITKQMICAGYKEGGIDACKGDSGGPLVCKHSGRWQLVGITSWGEGCARKEQPG 609
>sp|O15393|TMPS2_HUMAN Transmembrane protease, serine 2 precursor [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain] Length = 492 Score = 99.0 bits (245), Expect = 2e-20 Identities = 77/277 (27%), Positives = 115/277 (41%) Frame = +1 Query: 13 LSTITLAFKTNAKIMKRVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTKGNIS 192 L I N+ R++GGE A P + W VS+ +++ CG SI+ + Sbjct: 239 LRCIACGVNLNSSRQSRIVGGESALPGAWPWQVSL-----HVQNVHVCGGSIITPE---- 289 Query: 193 WLVTAAHCFYPSNKKKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSKAF 372 W+VTAAHC L P W A G+++ F++ +G G+ Sbjct: 290 WIVTAAHCVEKP-------LNNPWHWTAFAGILRQSFMF--YGAGY-------------- 326 Query: 373 RKLFQSKQNEVVTTGDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTIDMP 552 ++ +I HP Y S DIALMKL PL L V + +P Sbjct: 327 -----------------QVEKVISHPNYDSKTKNNDIALMKLQKPLTFNDL--VKPVCLP 367 Query: 553 DIPKGVSWPEVDSECVVVGWGCTKAAGVPNNIAKLAKLKVWDNQKCIDAFVAPIDLNPTS 732 + P + PE C + GWG T+ G + + AK+ + + Q+C +V + P + Sbjct: 368 N-PGMMLQPE--QLCWISGWGATEEKGKTSEVLNAAKVLLIETQRCNSRYVYDNLITP-A 423 Query: 733 EFCAGYLKKKYGLCPGDSGSGLACQYNGKWTFAGVVS 843 CAG+L+ C GDSG L N W G S Sbjct: 424 MICAGFLQGNVDSCQGDSGGPLVTSKNNIWWLIGDTS 460
>sp|P26262|KLKB1_MOUSE Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain] Length = 638 Score = 96.7 bits (239), Expect = 8e-20 Identities = 72/265 (27%), Positives = 113/265 (42%) Frame = +1 Query: 49 KIMKRVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTKGNISWLVTAAHCFYPS 228 KI R++GG A ++ W VS++ V + + CG SI+ + W++TAAHCF Sbjct: 386 KINARIVGGTNASLGEWPWQVSLQ--VKLVSQTHLCGGSIIGRQ----WVLTAAHCFD-- 437 Query: 229 NKKKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSKAFRKLFQSKQNEVV 408 + P++W G++ + I + Sbjct: 438 ------GIPYPDVWRIYGGILSLSEITKE------------------------------- 460 Query: 409 TTGDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTIDMPDIPKGVSWPEVD 588 T IK +IIH YK + YDIAL+KL PL + + + P + Sbjct: 461 -TPSSRIKELIIHQEYKVSEGNYDIALIKLQTPLNYTEFQKPICL-----PSKADTNTIY 514 Query: 589 SECVVVGWGCTKAAGVPNNIAKLAKLKVWDNQKCIDAFVAPIDLNPTSEFCAGYLKKKYG 768 + C V GWG TK G NI + A + + N++C + + +N CAGY + Sbjct: 515 TNCWVTGWGYTKEQGETQNILQKATIPLVPNEECQKKYRDYV-INKQM-ICAGYKEGGTD 572 Query: 769 LCPGDSGSGLACQYNGKWTFAGVVS 843 C GDSG L C+++G+W G+ S Sbjct: 573 ACKGDSGGPLVCKHSGRWQLVGITS 597
>sp|Q9JIQ8|TMPS2_MOUSE Transmembrane protease, serine 2 (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain] Length = 490 Score = 90.9 bits (224), Expect = 4e-18 Identities = 70/267 (26%), Positives = 106/267 (39%) Frame = +1 Query: 43 NAKIMKRVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTKGNISWLVTAAHCFY 222 + K R++GG A P + W VS+ ++ CG SI+ + W+VTAAHC Sbjct: 247 SVKRQSRIVGGLNASPGDWPWQVSL-----HVQGVHVCGGSIITPE----WIVTAAHCVE 297 Query: 223 PSNKKKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSKAFRKLFQSKQNE 402 L+ P W A G+++ +F +++ Sbjct: 298 EP-------LSGPRYWTAFAGILRQSL-------------------------MFYGSRHQ 325 Query: 403 VVTTGDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTIDMPDIPKGVSWPE 582 V + +I HP Y S DIALMKL PL L + +P + Sbjct: 326 V--------EKVISHPNYDSKTKNNDIALMKLQTPLAFNDL-----VKPVCLPNPGMMLD 372 Query: 583 VDSECVVVGWGCTKAAGVPNNIAKLAKLKVWDNQKCIDAFVAPIDLNPTSEFCAGYLKKK 762 +D EC + GWG T G +++ A + + + KC ++ + P CAG+L+ Sbjct: 373 LDQECWISGWGATYEKGKTSDVLNAAMVPLIEPSKCNSKYIYNNLITPAM-ICAGFLQGS 431 Query: 763 YGLCPGDSGSGLACQYNGKWTFAGVVS 843 C GDSG L NG W G S Sbjct: 432 VDSCQGDSGGPLVTLKNGIWWLIGDTS 458
>sp|Q7Z410|TMPS9_HUMAN Transmembrane protease, serine 9 (Polyserase-1) (Polyserase-I) (Polyserine protease-1) [Contains: Serase-1; Serase-2; Serase-3] Length = 1059 Score = 87.4 bits (215), Expect = 5e-17 Identities = 72/275 (26%), Positives = 115/275 (41%), Gaps = 6/275 (2%) Frame = +1 Query: 61 RVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTKGNISWLVTAAHCFYPSNKKK 240 R++GG EA P +F W S++ + + CGA+I+ N WLV+AAHCF Sbjct: 202 RIVGGMEASPGEFPWQASLRENKEHF-----CGAAII----NARWLVSAAHCFN------ 246 Query: 241 TFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSKAFRKLFQSKQNEVVTTGD 420 + P W A VG T+ S +T Sbjct: 247 --EFQDPTKWVAYVGA---------------------TYLS-----------GSEASTVR 272 Query: 421 IHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTIDMPDIPKGVSWPEVDSECV 600 + I+ HP Y ++ ++D+A+++L +PLP + I +P +C+ Sbjct: 273 AQVVQIVKHPLYNADTADFDVAVLELTSPLPFGR-----HIQPVCLPAATHIFPPSKKCL 327 Query: 601 VVGWGCTKAAG-VPNNIAKLAKLKVWDNQKCIDAFVAPIDLNPTSEFCAGYLKKKYGLCP 777 + GWG K V + + A +++ D C + + CAGYL K C Sbjct: 328 ISGWGYLKEDFLVKPEVLQKATVELLDQALCASLYGHSLT---DRMVCAGYLDGKVDSCQ 384 Query: 778 GDSGSGLACQY-NGKWTFAGVVS----AAHSKEPG 867 GDSG L C+ +G++ AG+VS A ++ PG Sbjct: 385 GDSGGPLVCEEPSGRFFLAGIVSWGIGCAEARRPG 419
Score = 74.7 bits (182), Expect = 3e-13
Identities = 74/302 (24%), Positives = 124/302 (41%), Gaps = 17/302 (5%)
Frame = +1
Query: 13 LSTITLAFKTNAKIMK-----------RVIGGEEAPPDKFRWAVSIKGDVDELKSSGACG 159
LST+ L + T K+ + RV+GG A + W VS+K S CG
Sbjct: 476 LSTVPLDWVTVPKLQECGARPAMEKPTRVVGGFGAASGEVPWQVSLKEG-----SRHFCG 530
Query: 160 ASILKTKGNISWLVTAAHCFYPSNKKKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTN 339
A+++ + WL++AAHCF + TK E A +G + + +
Sbjct: 531 ATVVGDR----WLLSAAHCF---------NHTKVEQVRAHLGTASLLGLGGSPVK----- 572
Query: 340 NKLITFFSKAFRKLFQSKQNEVVTTGDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIE 519
I ++ +++HP Y L++D+A+++L +PL
Sbjct: 573 ---------------------------IGLRRVVLHPLYNPGILDFDLAVLELASPLAFN 605
Query: 520 QLKGVNTIDMPDIPKGVSWPEVDSECVVVGWGCTKAAGVPN-NIAKLAKLKVWDNQKCID 696
+ I +P + V +C++ GWG T+ + + A + + D + C
Sbjct: 606 KY-----IQPVCLPLAIQKFPVGRKCMISGWGNTQEGNATKPELLQKASVGIIDQKTCSV 660
Query: 697 AFVAPIDLNPTSEFCAGYLKKKYGLCPGDSGSGLAC-QYNGKWTFAGVVS----AAHSKE 861
+ + CAG+L+ K C GDSG LAC + G + AG+VS A K+
Sbjct: 661 LYNFSLT---DRMICAGFLEGKVDSCQGDSGGPLACEEAPGVFYLAGIVSWGIGCAQVKK 717
Query: 862 PG 867
PG
Sbjct: 718 PG 719
Score = 73.6 bits (179), Expect = 7e-13
Identities = 68/265 (25%), Positives = 113/265 (42%), Gaps = 2/265 (0%)
Frame = +1
Query: 55 MKRVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTKGNISWLVTAAHCFYPSNK 234
+ R++GG A ++ W VS+ E + CGA ++ + WL++AAHCF
Sbjct: 824 LTRIVGGSAAGRGEWPWQVSLWLRRREHR----CGAVLVAER----WLLSAAHCFDVYGD 875
Query: 235 KKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSKAFRKLFQSKQNEVVTT 414
P+ W A +G F S A +L +
Sbjct: 876 --------PKQWAAFLGT---------------------PFLSGAEGQLER--------- 897
Query: 415 GDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTIDMPD-IPKGVSWPEVDS 591
+ I HP+Y L+YD+AL++L P+ +L V I +P+ P+ P +
Sbjct: 898 ----VARIYKHPFYNLYTLDYDVALLELAGPVRRSRL--VRPICLPEPAPR----PPDGT 947
Query: 592 ECVVVGWGCTKAAGVPNNIAKLAKLKVWDNQKCIDAFVAPIDLNPTSEFCAGYLKKKYGL 771
CV+ GWG + G + A +++ Q C + P+ ++ + CAG+ +
Sbjct: 948 RCVITGWGSVREGGSMARQLQKAAVRLLSEQTCRRFY--PVQIS-SRMLCAGFPQGGVDS 1004
Query: 772 CPGDSGSGLAC-QYNGKWTFAGVVS 843
C GD+G LAC + +G+W GV S
Sbjct: 1005 CSGDAGGPLACREPSGRWVLTGVTS 1029
>sp|P03952|KLKB1_HUMAN Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain] Length = 638 Score = 85.9 bits (211), Expect = 1e-16 Identities = 77/278 (27%), Positives = 117/278 (42%), Gaps = 5/278 (1%) Frame = +1 Query: 49 KIMKRVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTKGNISWLVTAAHCFYPS 228 K R++GG + ++ W VS++ V CG S++ + W++TAAHCF Sbjct: 386 KTSTRIVGGTNSSWGEWPWQVSLQ--VKLTAQRHLCGGSLIGHQ----WVLTAAHCFD-- 437 Query: 229 NKKKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSKAFRKLFQSKQNEVV 408 L ++W G++ + I T FS+ Sbjct: 438 ------GLPLQDVWRIYSGILNLSDITKD------------TPFSQ-------------- 465 Query: 409 TTGDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTIDMPDIPKGVSWPEVD 588 IK IIIH YK + +DIAL+KL PL + + I +P KG + + Sbjct: 466 ------IKEIIIHQNYKVSEGNHDIALIKLQAPLNYTEFQ--KPICLPS--KGDT-STIY 514 Query: 589 SECVVVGWGCTKAAGVPNNIAKLAKLKVWDNQKCIDAFVAPIDLNPTSEF-CAGYLKKKY 765 + C V GWG +K G NI + + + N++C + D T CAGY + Sbjct: 515 TNCWVTGWGFSKEKGEIQNILQKVNIPLVTNEECQKRYQ---DYKITQRMVCAGYKEGGK 571 Query: 766 GLCPGDSGSGLACQYNGKWTFAGVVS----AAHSKEPG 867 C GDSG L C++NG W G+ S A ++PG Sbjct: 572 DACKGDSGGPLVCKHNGMWRLVGITSWGEGCARREQPG 609
>sp|P98074|ENTK_PIG Enteropeptidase precursor (Enterokinase) [Contains: Enteropeptidase non-catalytic mini chain; Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain] Length = 1034 Score = 83.2 bits (204), Expect = 9e-16 Identities = 72/270 (26%), Positives = 110/270 (40%), Gaps = 5/270 (1%) Frame = +1 Query: 49 KIMKRVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTKGNISWLVTAAHCFYPS 228 ++ +++GG ++ + W V++ + L CGAS++ WLV+AAHC Y Sbjct: 795 EVSPKIVGGNDSREGAWPWVVALYYNGQLL-----CGASLVSR----DWLVSAAHCVYGR 845 Query: 229 NKKKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSKAFRKLFQSKQNEVV 408 N +P W A +GL H LT+ +++T Sbjct: 846 N-------LEPSKWKAILGL---------HMTSNLTSPQIVTRL---------------- 873 Query: 409 TTGDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTIDMPD----IPKGVSW 576 I I+I+P+Y R + DIA+M L + + I +P+ P G Sbjct: 874 ------IDEIVINPHYNRRRKDSDIAMMHL--EFKVNYTDYIQPICLPEENQVFPPG--- 922 Query: 577 PEVDSECVVVGWGCTKAAGVPNNIAKLAKLKVWDNQKCIDAFVAPIDLNPTSEF-CAGYL 753 C + GWG G P +I + A + + N+KC + N T CAGY Sbjct: 923 ----RICSIAGWGKVIYQGSPADILQEADVPLLSNEKCQQQMP---EYNITENMMCAGYE 975 Query: 754 KKKYGLCPGDSGSGLACQYNGKWTFAGVVS 843 + C GDSG L C N +W AGV S Sbjct: 976 EGGIDSCQGDSGGPLMCLENNRWLLAGVTS 1005
>sp|P56677|ST14_MOUSE Suppressor of tumorigenicity 14 (Epithin) Length = 855 Score = 82.8 bits (203), Expect = 1e-15 Identities = 81/279 (29%), Positives = 116/279 (41%), Gaps = 10/279 (3%) Frame = +1 Query: 61 RVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTKGNISWLVTAAHCFYPSNKKK 240 RV+GG A ++ W VS+ L CGAS++ WLV+AAHCF K Sbjct: 614 RVVGGTNADEGEWPWQVSLHA----LGQGHLCGASLISP----DWLVSAAHCFQDDKNFK 665 Query: 241 TFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSKAFRKLFQSKQNEVVTTGD 420 D T +W A +GL+ QSK++ + Sbjct: 666 YSDYT---MWTAFLGLLD------------------------------QSKRS-ASGVQE 691 Query: 421 IHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTIDMPD----IPKGVS-WPEV 585 + +K II HP + +YDIAL++L +E V I +PD P G + W Sbjct: 692 LKLKRIITHPSFNDFTFDYDIALLEL--EKSVEYSTVVRPICLPDATHVFPAGKAIW--- 746 Query: 586 DSECVVVGWGCTKAAGVPNNIAKLAKLKVWDNQKCIDAFVAPIDLNPTSEFCAGYLKKKY 765 V GWG TK G I + +++V + C D + P + P C G+L Sbjct: 747 -----VTGWGHTKEGGTGALILQKGEIRVINQTTCED--LMPQQITPRM-MCVGFLSGGV 798 Query: 766 GLCPGDSGSGL-ACQYNGKWTFAGVVS----AAHSKEPG 867 C GDSG L + + +G+ AGVVS A +PG Sbjct: 799 DSCQGDSGGPLSSAEKDGRMFQAGVVSWGEGCAQRNKPG 837
>sp|Q9ER04|TMPS5_MOUSE Transmembrane protease, serine 5 (Spinesin) Length = 455 Score = 82.0 bits (201), Expect = 2e-15 Identities = 69/277 (24%), Positives = 105/277 (37%), Gaps = 5/277 (1%) Frame = +1 Query: 52 IMKRVIGGEEAPPDKFRWAVSIKGDVDELKSSGACGASILKTKGNISWLVTAAHCFYPSN 231 + R++GG+ ++ W S+ L S CGAS+L W+VTAAHC Y Sbjct: 214 LASRIVGGQAVASGRWPWQASVM-----LGSRHTCGASVLAPH----WVVTAAHCMY--- 261 Query: 232 KKKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSKAFRKLFQSKQNEVVT 411 +F L++ W GL+ + G Sbjct: 262 ---SFRLSRLSSWRVHAGLVSHGAVRQHQG------------------------------ 288 Query: 412 TGDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTIDMPDIPKGVSWPEVDS 591 ++ II HP Y + +YD+AL++L P+ +T+D +P + S Sbjct: 289 ---TMVEKIIPHPLYSAQNHDYDVALLQLRTPINFS-----DTVDAVCLPAKEQYFPWGS 340 Query: 592 ECVVVGWGCTKAAGV-PNNIAKLAKLKVWDNQKCIDAFVAPIDLNPTSEFCAGYLKKKYG 768 +C V GWG T + ++ + + + C + + L CAGYL + Sbjct: 341 QCWVSGWGHTDPSHTHSSDTLQDTMVPLLSTHLCNSSCMYSGALTHRM-LCAGYLDGRAD 399 Query: 769 LCPGDSGSGLACQYNGKWTFAGVVS----AAHSKEPG 867 C GDSG L C W GVVS A PG Sbjct: 400 ACQGDSGGPLVCPSGDTWHLVGVVSWGRGCAEPNRPG 436
>sp|P23578|ACRO_MOUSE Acrosin precursor [Contains: Acrosin light chain; Acrosin heavy chain] Length = 436 Score = 81.6 bits (200), Expect = 3e-15 Identities = 79/294 (26%), Positives = 120/294 (40%), Gaps = 8/294 (2%) Frame = +1 Query: 10 CLSTITLAFKTNAKIMKRVIGGEEAPPDKFRWAVSIKGDVDE-LKSSGACGASILKTKGN 186 C L F+ N++ R++ G+ A + W VS++ + ACG S+L N Sbjct: 25 CDGPCGLRFRQNSQAGTRIVSGQSAQLGAWPWMVSLQIFTSHNSRRYHACGGSLL----N 80 Query: 187 ISWLVTAAHCFYPSNKKKTFDLTKPELWHARVGLMKVDFIYNKHGRGFLTNNKLITFFSK 366 W++TAAHCF NKKK +D W G ++++ NK + Sbjct: 81 SHWVLTAAHCF--DNKKKVYD------WRLVFGAQEIEYGRNKPVK-------------- 118 Query: 367 AFRKLFQSKQNEVVTTGDIHIKNIIIHPYYKSNRLEYDIALMKLINPLPIEQLKGVNTID 546 + +Q +++ I+IH Y DIAL+K+ P+ G Sbjct: 119 ------EPQQER-------YVQKIVIHEKYNVVTEGNDIALLKITPPVTCGNFIG--PCC 163 Query: 547 MPDIPKGVSWPEVDSECVVVGWGCTKA-AGVPNNIAKLAKLKVWDNQKCIDAFVAPIDLN 723 +P G P++ C V GWG K A P+ + A++ + D C + Sbjct: 164 LPHFKAGP--PQIPHTCYVTGWGYIKEKAPRPSPVLMEARVDLIDLDLCNSTQWYNGRVT 221 Query: 724 PTSEFCAGYLKKKYGLCPGDSGSGLACQYNGKWTFAGV------VSAAHSKEPG 867 T+ CAGY + K C GDSG L C+ N F V V A +K PG Sbjct: 222 STN-VCAGYPEGKIDTCQGDSGGPLMCRDNVDSPFVVVGITSWGVGCARAKRPG 274
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 98,995,733
Number of Sequences: 369166
Number of extensions: 2064252
Number of successful extensions: 6432
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5441
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6005
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 8550075140
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail