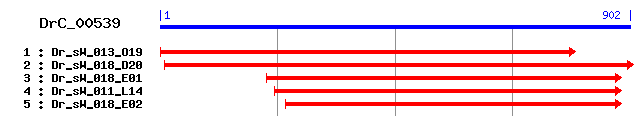
DrC_00539
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00539 (902 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P07207|NOTCH_DROME Neurogenic locus Notch protein precursor 51 5e-06 sp|P10039|TENA_CHICK Tenascin precursor (TN) (Hexabrachion)... 49 1e-05 sp|P10968|AGI1_WHEAT Agglutinin isolectin 1 precursor (WGA1... 48 4e-05 sp|P24821|TENA_HUMAN Tenascin precursor (TN) (Hexabrachion)... 47 7e-05 sp|P22105|TENX_HUMAN Tenascin-X precursor (TN-X) (Hexabrach... 46 2e-04 sp|P41951|PQN25_CAEEL Glutamine/asparagine-rich protein pqn-25 45 2e-04 sp|Q29116|TENA_PIG Tenascin precursor (TN) (Hexabrachion) (... 45 4e-04 sp|P02876|AGI2_WHEAT Agglutinin isolectin 2 precursor (WGA2... 44 6e-04 sp|P10969|AGI3_WHEAT Agglutinin isolectin 3 precursor (WGA3) 44 6e-04 sp|P10040|CRB_DROME Crumbs protein precursor (95F) 43 0.001
>sp|P07207|NOTCH_DROME Neurogenic locus Notch protein precursor Length = 2703 Score = 50.8 bits (120), Expect = 5e-06 Identities = 61/220 (27%), Positives = 82/220 (37%), Gaps = 15/220 (6%) Frame = +1 Query: 100 DSKSSNLVGGGGACRRHVECFSGLCVKRDTGKVGFCSKDSSVIKLIGGGGIC--KNNGQC 273 D SSN GG C + FS C+ TG+ + D V G GG C K NG Sbjct: 793 DECSSNPCQHGGTCYDKLNAFSCQCMPGYTGQKCETNIDDCVTNPCGNGGTCIDKVNG-- 850 Query: 274 YSRYCYKVDAETKICSKDSKFKGKVGGGGPCKNDFFC--ASNF------CHEINGIKYCV 429 YK + +D + K CKN+ C +SNF C +YC Sbjct: 851 -----YKCVCKVPFTGRDCESKMDPCASNRCKNEAKCTPSSNFLDFSCTCKLGYTGRYCD 905 Query: 430 QESKQ--KIGYIGEGALCKNHHSCYSGYCVKVGNEG-FCA--KESMKSEKVGGGGPCVRD 594 ++ + GA C N Y C K G EG CA + S GG C+ Sbjct: 906 EDIDECSLSSPCRNGASCLNVPGSYRCLCTK-GYEGRDCAINTDDCASFPCQNGGTCLDG 964 Query: 595 WQCYSRYCFNNGKLRYCAKDGKQFGEVGISGPCRKNKDCS 714 YS C + ++C D + +S PC+ CS Sbjct: 965 IGDYSCLCVDGFDGKHCETDINEC----LSQPCQNGATCS 1000
Score = 36.6 bits (83), Expect = 0.099
Identities = 40/145 (27%), Positives = 49/145 (33%), Gaps = 11/145 (7%)
Frame = +1
Query: 112 SNLVGGGGACRRHVECFSGLCVKRDTGKVGFCSKDSSVIKLIGGGGICKNNGQCYSRYCY 291
SN GG C + +C TGK GGIC++NG Y C
Sbjct: 223 SNPCKYGGTCVNTHGSYQCMCPTGYTGKDCDTKYKPCSPSPCQNGGICRSNGLSYECKCP 282
Query: 292 KVDAETKICSK--DSKFKGKVGGGGPC---KNDFFC------ASNFCHEINGIKYCVQES 438
K E K C + D GG C +D+ C FC + + C Q
Sbjct: 283 K-GFEGKNCEQNYDDCLGHLCQNGGTCIDGISDYTCRCPPNFTGRFCQD--DVDECAQRD 339
Query: 439 KQKIGYIGEGALCKNHHSCYSGYCV 513
GA C N H YS CV
Sbjct: 340 HP---VCQNGATCTNTHGSYSCICV 361
Score = 33.1 bits (74), Expect = 1.1
Identities = 59/256 (23%), Positives = 85/256 (33%), Gaps = 38/256 (14%)
Frame = +1
Query: 43 KMCFSGVCVLLGNMQVC-------------AKDSKSSNLVGGGGACRRHVECFSGLCVKR 183
++C +G C GN VC D S GG CR + + C +
Sbjct: 1153 QLCNNGTCKDYGNSHVCYCSQGYAGSYCQKEIDECQSQPCQNGGTCRDLIGAYECQCRQG 1212
Query: 184 DTGKVGFCSKDSSVIKLIGGGGICKNNGQCYSRYC-----------YKVDAETKICSKDS 330
G+ + D GG C + +S C K D + C +
Sbjct: 1213 FQGQNCELNIDDCAPNPCQNGGTCHDRVMNFSCSCPPGTMGIICEINKDDCKPGACHNNG 1272
Query: 331 KFKGKVGG----------GGPCKNDF-FCASNFCHEINGIKYCVQESKQKIGYIGEGALC 477
+VGG G C+ D C SN C G CVQ L
Sbjct: 1273 SCIDRVGGFECVCQPGFVGARCEGDINECLSNPCSNA-GTLDCVQ-------------LV 1318
Query: 478 KNHH-SCYSGYCVK--VGNEGFCAKESMKSEKVGGGGPCVRDWQCYSRYCFNNGKLRYCA 648
N+H +C G+ + FCA+ ++ GG C + C NNG +
Sbjct: 1319 NNYHCNCRPGHMGRHCEHKVDFCAQSPCQN-----GGNCNIRQSGHHCIC-NNG---FYG 1369
Query: 649 KDGKQFGEVGISGPCR 696
K+ + G+ S PCR
Sbjct: 1370 KNCELSGQDCDSNPCR 1385
>sp|P10039|TENA_CHICK Tenascin precursor (TN) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Miotendinous antigen) (Glioma-associated-extracellular matrix antigen) (GP 150-225) Length = 1808 Score = 49.3 bits (116), Expect = 1e-05 Identities = 65/255 (25%), Positives = 96/255 (37%), Gaps = 22/255 (8%) Frame = +1 Query: 31 CTEDKMCFSGVCV----LLGN---MQVCAKDSKSSNLVGGGGACRRHVECFSGLCVKRDT 189 C + C +G+CV +G+ + C KD C C G CV + Sbjct: 351 CNGNGRCENGLCVCHEGFVGDDCSQKRCPKD------------CNNRGHCVDGRCVCHE- 397 Query: 190 GKVGFCSKDSSVIKLIGGGGICKNNGQCYSRYCYKVDAETKICSKDSKFKGKVGGGGPCK 369 G+ +D ++ C N G+C + C +C D F G+ G C Sbjct: 398 ---GYLGEDCGELRCPND---CHNRGRCINGQC--------VC--DEGFIGEDCGELRCP 441 Query: 370 NDFFCASNFCHE----INGIKYCVQESKQKIGYIGE--GAL-----CKNHHSCYSGYCVK 516 ND CH +NG C + G+IGE G L C +H C +G CV Sbjct: 442 ND-------CHNRGRCVNGQCECHE------GFIGEDCGELRCPNDCNSHGRCVNGQCVC 488 Query: 517 VGNEGFCAKESMKSEKVGGGGPCVRDWQCYSRYCFNNGKLRYCAKD----GKQFGEVGIS 684 +EG+ ++ G C D C++R G+ C D G+ GE+ Sbjct: 489 --DEGYTGEDC-------GELRCPND--CHNRGRCVEGR---CVCDNGFMGEDCGELSCP 534 Query: 685 GPCRKNKDCSSGRCV 729 C ++ C GRCV Sbjct: 535 NDCHQHGRCVDGRCV 549
Score = 32.7 bits (73), Expect = 1.4
Identities = 58/236 (24%), Positives = 87/236 (36%), Gaps = 19/236 (8%)
Frame = +1
Query: 136 ACRRHVECFS-GLCVK-RDTGKVGFCSKDSSVIKLIGGGGICKNNGQCYSRYCYKVDAET 309
AC R+ C + GLCV+ + + GF +D S C + G+C C + T
Sbjct: 191 ACPRN--CLNRGLCVRGKCICEEGFTGEDCSQAACPSD---CNDQGKCVDGVCVCFEGYT 245
Query: 310 ------KICSKDSKFKGKVGGGGPCKNDFFCASNFCHEINGIKYCVQESKQKIGYIGEGA 471
++C G+ GG C CHE G+ GE
Sbjct: 246 GPDCGEELCPHGCGIHGRCVGGR-------CV---CHE---------------GFTGEDC 280
Query: 472 ---LCKNHHSCYS-GYCVK---VGNEGFCAKESMKSEKVGGGGPCVRDWQCYSRYCFNNG 630
LC N+ C++ G CV V +EG+ ++ G + C+ R NG
Sbjct: 281 NEPLCPNN--CHNRGRCVDNECVCDEGYTGEDC---------GELICPNDCFDRGRCING 329
Query: 631 KLRYCAKD--GKQFGEVGISGPCRKNKDCSSGRCVSMKTKLQDGTESK--VKVCQN 786
+C + G+ GE+ C N C +G CV + + D K K C N
Sbjct: 330 TC-FCEEGYTGEDCGELTCPNNCNGNGRCENGLCVCHEGFVGDDCSQKRCPKDCNN 384
>sp|P10968|AGI1_WHEAT Agglutinin isolectin 1 precursor (WGA1) (Isolectin A) Length = 212 Score = 47.8 bits (112), Expect = 4e-05 Identities = 51/169 (30%), Positives = 60/169 (35%), Gaps = 7/169 (4%) Frame = +1 Query: 241 GGGICKNNGQCYSRYCYKVDAETKICSKDSKFKGKVGGGGPCKNDFFCASNFCHEINGIK 420 GG C NN QC S+Y Y C +++ G GGPC+ D C S Sbjct: 77 GGATCTNN-QCCSQYGY--------CGFGAEYCGAGCQGGPCRADIKCGSQ--------- 118 Query: 421 YCVQESKQKIGYIGEGALCKNHHSCYSGYCVKVGNEGFCAKESMKSEKVGGG---GPCVR 591 G LC N+ C GFC + SE GGG G C Sbjct: 119 -------------AGGKLCPNNLCC--------SQWGFC---GLGSEFCGGGCQSGACST 154 Query: 592 DWQC----YSRYCFNNGKLRYCAKDGKQFGEVGISGPCRKNKDCSSGRC 726 D C R C NN YC ++G GI GP C SG C Sbjct: 155 DKPCGKDAGGRVCTNN----YCC---SKWGSCGI-GPGYCGAGCQSGGC 195
>sp|P24821|TENA_HUMAN Tenascin precursor (TN) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Miotendinous antigen) (Glioma-associated-extracellular matrix antigen) (GP 150-225) (Tenascin-C) (TN-C) Length = 2201 Score = 47.0 bits (110), Expect = 7e-05 Identities = 63/244 (25%), Positives = 85/244 (34%), Gaps = 11/244 (4%) Frame = +1 Query: 31 CTEDKMCFSGVCVLLGNMQVCAKDSKSSNLVGGGGACRRHVECFSGLCVKRDTGKVGFCS 210 C + C +GVC+ + S + H C GLCV D GF Sbjct: 225 CNDQGKCVNGVCICFEGYA----GADCSREICPVPCSEEHGTCVDGLCVCHD----GFAG 276 Query: 211 KDSSVIKLIGGGGICKNNGQCYSRYCYKVDAETKICSKDSKFKGKVGGGGPCKNDFFCAS 390 D + + C N G+C C +C D F G+ C ND F Sbjct: 277 DDCNKPLCLNN---CYNRGRCVENEC--------VC--DEGFTGEDCSELICPNDCFDRG 323 Query: 391 NFCHEINGIKYCVQESKQKIGYIGEGA-------LCKNHHSCYSGYCVKVGNEGFCAKES 549 ING YC + G+ GE C C G C V +EGF + Sbjct: 324 RC---INGTCYCEE------GFTGEDCGKPTCPHACHTQGRCEEGQC--VCDEGFAGLDC 372 Query: 550 MKSEKVGGGGPCVRDWQCYSRYCFNNGKLRYCAKD----GKQFGEVGISGPCRKNKDCSS 717 SEK C D C++R +G+ C D G GE+ C + C + Sbjct: 373 --SEK-----RCPAD--CHNRGRCVDGR---CECDDGFTGADCGELKCPNGCSGHGRCVN 420 Query: 718 GRCV 729 G+CV Sbjct: 421 GQCV 424
Score = 40.0 bits (92), Expect = 0.009
Identities = 59/234 (25%), Positives = 78/234 (33%), Gaps = 23/234 (9%)
Frame = +1
Query: 136 ACRRHVECFSGLCVKRDTGKVGFCSKDSSVIKLIGGGGICKNNGQCYSRYCYKVDAETKI 315
AC C G CV + GF D S + C N G+C C D T
Sbjct: 349 ACHTQGRCEEGQCVCDE----GFAGLDCSEKRCPAD---CHNRGRCVDGRCECDDGFTGA 401
Query: 316 CSKDSKFKGKVGGGGPCKNDFFCASNFCHEINGIKYCVQESKQKIGYIGEGAL------- 474
+ K G G C +NG C + GY GE
Sbjct: 402 DCGELKCPNGCSGHGRC-------------VNGQCVCDE------GYTGEDCSQLRCPND 442
Query: 475 CKNHHSCYSGYCV-KVGNEGF-CAKESMKSEKVGGG----GPCVRD-----WQCYSRYCF 621
C + C G CV + G +G+ C+ S ++ G G CV D C R C
Sbjct: 443 CHSRGRCVEGKCVCEQGFKGYDCSDMSCPNDCHQHGRCVNGMCVCDDGYTGEDCRDRQCP 502
Query: 622 NNGKLRYCAKDGKQFGEVGISGPCRKNKDC-----SSGRCVSMKTKLQDGTESK 768
+ R DG+ E G +GP C GRCV+ + +G K
Sbjct: 503 RDCSNRGLCVDGQCVCEDGFTGPDCAELSCPNDCHGQGRCVNGQCVCHEGFMGK 556
>sp|P22105|TENX_HUMAN Tenascin-X precursor (TN-X) (Hexabrachion-like protein) Length = 4289 Score = 45.8 bits (107), Expect = 2e-04 Identities = 66/284 (23%), Positives = 100/284 (35%), Gaps = 27/284 (9%) Frame = +1 Query: 16 GGGGQCTEDK-MCFSGVCVLLGNMQVCAKDSKSSNLVGGGGACRRHVECFSGLCVKRDTG 192 G GG+C + + +C+ G + + C +D CR C G C+ DTG Sbjct: 347 GEGGRCVDGRCVCWPGYTGEDCSTRTCPRD------------CRGRGRCEDGECIC-DTG 393 Query: 193 KVGFCSKDSSVIKLIGGGGICKNNGQCYSRYCY------KVDAETKICSKDSKFKGKVGG 354 G D ++ G C G+C C D ++ C +D + G Sbjct: 394 YSG----DDCGVRSCPGD--CNQRGRCEDGRCVCWPGYTGTDCGSRACPRDCR------G 441 Query: 355 GGPCKNDF-FCASNFCHEINGIKYCVQESKQKIGYIGEGALCKNHHSCYSGYCVKVGNEG 531 G C+N C + + E G++ C + + G G C+ GY G + Sbjct: 442 RGRCENGVCVCNAGYSGEDCGVRSCPGDCR------GRGRCESGRCMCWPGY---TGRD- 491 Query: 532 FCAKESMKSEKVGGG----GPCVRD-----WQCYSRYCFNNGKLRYCAKDGKQFGEVGIS 684 C + + G G G CV + C SR C + + +DG + G S Sbjct: 492 -CGTRACPGDCRGRGRCVDGRCVCNPGFTGEDCGSRRCPGDCRGHGLCEDGVCVCDAGYS 550 Query: 685 GP----------CRKNKDCSSGRCVSMKTKLQDGTESKVKVCQN 786 G CR C GRCV G + V+ C N Sbjct: 551 GEDCSTRSCPGGCRGRGQCLDGRCVCEDG--YSGEDCGVRQCPN 592
Score = 43.5 bits (101), Expect = 8e-04
Identities = 68/264 (25%), Positives = 82/264 (31%), Gaps = 29/264 (10%)
Frame = +1
Query: 25 GQCTEDKMCFSGVCVLLGNMQVCAKDSKSSNLVGGGGACRRHVECFSGLCVKRDTGKVGF 204
G C C G CV N +D S G CR H C G+CV G+
Sbjct: 499 GDCRGRGRCVDGRCVC--NPGFTGEDCGSRRCPGD---CRGHGLCEDGVCVC----DAGY 549
Query: 205 CSKDSSVIKLIGGGGICKNNGQCYSRYCYKVDAET------KICSKDSKFKGKVGGGGPC 366
+D S GG C+ GQC C D + + C D G V G C
Sbjct: 550 SGEDCSTRSCPGG---CRGRGQCLDGRCVCEDGYSGEDCGVRQCPNDCSQHG-VCQDGVC 605
Query: 367 KNDFFCASNFCHEINGIKYCVQESKQK-----------IGYIGEG-------ALCKNHHS 492
C + E I+ C + GY G A C+
Sbjct: 606 ----ICWEGYVSEDCSIRTCPSNCHGRGRCEEGRCLCDPGYTGPTCATRMCPADCRGRGR 661
Query: 493 CYSGYC---VKVGNEGFCAKESMKSEKVGGGGP--CVRDWQCYSRYCFNNGKLRYCAKDG 657
C G C V G E +E S GG GP R QC C + CA
Sbjct: 662 CVQGVCLCHVGYGGEDCGQEEPPASACPGGCGpreLCRAGQCV---CVEGFRGPDCAIQT 718
Query: 658 KQFGEVGISGPCRKNKDCSSGRCV 729
G CR +C G CV
Sbjct: 719 -------CPGDCRGRGECHDGSCV 735
Score = 42.4 bits (98), Expect = 0.002
Identities = 59/252 (23%), Positives = 90/252 (35%), Gaps = 19/252 (7%)
Frame = +1
Query: 31 CTEDKMCFSGVCVLLGNMQVCAKDSKSSNLVGGGGACRRHVECFSGLCVKRDTGKVGFCS 210
C++ C +G CV N +D + G C + C G CV D G G
Sbjct: 284 CSQRGRCENGRCVC--NPGYTGEDCGVRSCPRG---CSQRGRCKDGRCVC-DPGYTGEDC 337
Query: 211 KDSSVIKLIGGGGICKNNGQCYSRYCYK-VDAETKICSKDSKFKGKVGGGGPCKNDFFCA 387
S G GG C + G+C Y D T+ C +D + +G+ G + C
Sbjct: 338 GTRSCPWDCGEGGRCVD-GRCVCWPGYTGEDCSTRTCPRDCRGRGRCEDG-----ECICD 391
Query: 388 SNFCHEINGIKYCVQESKQK-----------IGYIGEGAL-------CKNHHSCYSGYCV 513
+ + + G++ C + Q+ GY G C+ C +G CV
Sbjct: 392 TGYSGDDCGVRSCPGDCNQRGRCEDGRCVCWPGYTGTDCGSRACPRDCRGRGRCENGVCV 451
Query: 514 KVGNEGFCAKESMKSEKVGGGGPCVRDWQCYSRYCFNNGKLRYCAKDGKQFGEVGISGPC 693
N G+ ++ G C D + R C + + + G+ G G C
Sbjct: 452 C--NAGYSGEDC-------GVRSCPGDCRGRGR-CESGRCMCWPGYTGRDCGTRACPGDC 501
Query: 694 RKNKDCSSGRCV 729
R C GRCV
Sbjct: 502 RGRGRCVDGRCV 513
>sp|P41951|PQN25_CAEEL Glutamine/asparagine-rich protein pqn-25 Length = 672 Score = 45.4 bits (106), Expect = 2e-04 Identities = 67/329 (20%), Positives = 105/329 (31%), Gaps = 70/329 (21%) Frame = +1 Query: 28 QCTEDKMCFSGVCVLLGNMQVCAKDSKSSNLVG-------------------------GG 132 QC C +G C N C+ + + L G GG Sbjct: 131 QCASGYTCNNGACCPNTNSNTCSSNGNNGCLAGQTMVNGQCYNSVNIGSACQSTQQCLGG 190 Query: 133 GACRRHV-ECFSGL------CVKRD--TGKVGFCSKDSSVIKLIGGGGICKNNGQCYSR- 282 C+ ++ +C+SG CV + ++G S +S I L G C+ + QC Sbjct: 191 SQCQNNICQCYSGYVNVNQQCVISNGLNCQLGTVSYNSQCITLASPGQNCQTSSQCIDNS 250 Query: 283 ----------------YCYKVDAETKICSKDSKFKGK-------VG----GGGPCKNDFF 381 Y Y V + IC + VG C Sbjct: 251 VCMNQMCTCNNNYRLVYGYCVPITSSICQQTQTLVNNQCVLLSIVGETCIANQQCVGGAM 310 Query: 382 CASNFCHEINGIK----YCVQESKQKIGYIGEGALCKNHHSCYSGYC---VKVGNEGFCA 540 C S C NG YC+ S C ++ +G C V+VG G C+ Sbjct: 311 CNSGTCQCTNGATAMYGYCISSSSSS---------CNSNQVSINGMCYNTVQVG--GSCS 359 Query: 541 KESMKSEKVGGGGPCVRDWQCYSRYCFNNGKLRYCAKDGKQFGEVGISGPCRKNKDC-SS 717 S++ C + C S +C + + + V I C ++ C S+ Sbjct: 360 ----FSQQCLNNAVCTNN-ICVSTFCSVSCSTNQVCISNQCYNYVSIGSQCVGSQQCLSN 414 Query: 718 GRCVSMKTKLQDGTESKVKVCQNPKKKDN 804 +C+S + GT+ VC +N Sbjct: 415 SQCISSICQCPQGTQQSNGVCTGNNNNNN 443
Score = 40.4 bits (93), Expect = 0.007
Identities = 53/272 (19%), Positives = 90/272 (33%), Gaps = 10/272 (3%)
Frame = +1
Query: 19 GGGQCTEDKMCFSGVCVLLGNMQVCAKDSKSSNLVGGG-----GACRRHVECFSGLCVKR 183
G QC + C S +C C + ++ SN V G C+ + + C
Sbjct: 407 GSQQCLSNSQCISSIC-------QCPQGTQQSNGVCTGNNNNNNQCQPNQVLINNQCY-- 457
Query: 184 DTGKVGF-CSKDSSVIKLIGGGGICKNNGQCYSRYCYKVDAETKICSKDSKFKGKVGGGG 360
+T +GF C C N QC + C T + + +G G G
Sbjct: 458 NTVSIGFQCQFPQQ----------CLGNSQCMNSMCQCPTGSTNV---NGYCQG--GSNG 502
Query: 361 PCKNDFFCASNFCHEINGIKYCVQESKQKIG---YIGEGALCKNHHSCYSGYCVKVGNEG 531
C ++ +N C+ I + Q ++Q +G + C + S +GYC
Sbjct: 503 QCNSNQVLINNQCYNTVSIGFQCQFAQQCLGNSQCLNSICQCPSGSSNVNGYC------- 555
Query: 532 FCAKESMKSEKVGGGGPCVRDWQCYSRYCFNNGKLRYCAKDGKQFGEVGISGPCRKNKDC 711
+ G G C + Y+ C+N V I C+ + C
Sbjct: 556 ----------QGGSNGQCNSNQVYYNNQCYNT---------------VPIGSQCQITQQC 590
Query: 712 -SSGRCVSMKTKLQDGTESKVKVCQNPKKKDN 804
+ +C++ + GT + C N
Sbjct: 591 LGNSQCMNSFCQCPSGTTNVNNFCTTSSSSSN 622
Score = 30.0 bits (66), Expect = 9.2
Identities = 30/118 (25%), Positives = 48/118 (40%), Gaps = 12/118 (10%)
Frame = +1
Query: 466 GALCKNHHSCYSGYCVKVGNEGFCAKESMKSEKVGGGGP---CVRDWQCYSRYCFNNG-- 630
G +C+ + S Y+ Y C S S +V G C + QC S Y NNG
Sbjct: 87 GTICQ-YASTYNNYICCYSTNTQCGSNS--SPQVSASGQVVTCSTNTQCASGYTCNNGAC 143
Query: 631 ----KLRYCAKDGKQ---FGEVGISGPCRKNKDCSSGRCVSMKTKLQDGTESKVKVCQ 783
C+ +G G+ ++G C + + S C S + L G++ + +CQ
Sbjct: 144 CPNTNSNTCSSNGNNGCLAGQTMVNGQCYNSVNIGSA-CQSTQQCL-GGSQCQNNICQ 199
>sp|Q29116|TENA_PIG Tenascin precursor (TN) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Miotendinous antigen) (Glioma-associated-extracellular matrix antigen) (GP 150-225) (Tenascin-C) (TN-C) (P230) Length = 1746 Score = 44.7 bits (104), Expect = 4e-04 Identities = 71/288 (24%), Positives = 96/288 (33%), Gaps = 32/288 (11%) Frame = +1 Query: 19 GGGQCTEDK-MCFSGVCVLLGNMQVCAKDSKSSNLVGGGGACRRHVECFSGLCVKRDTGK 195 G G+C E + +C G + + C D C C G C D Sbjct: 352 GRGRCEEGQCVCDEGFAGADCSERRCPSD------------CHNRGRCLDGRCECDD--- 396 Query: 196 VGFCSKDSSVIKLIGGGGICKNNGQCYSRYCYKVDAETKI------CSKDSKFKGK-VGG 354 GF +D ++ GG C +G+C + C + T C D +G+ V G Sbjct: 397 -GFEGEDCGELRCPGG---CSGHGRCVNGQCVCDEGRTGEDCSQLRCPNDCHGRGRCVQG 452 Query: 355 GGPCKNDF-------FCASNFCHE----INGIKYCVQESKQKIGYIGEGAL-------CK 480 C++ F + CH+ +NG+ C GY GE C Sbjct: 453 RCECEHGFQGYDCSEMSCPHDCHQHGRCVNGMCVCDD------GYTGEDCRELRCPGDCS 506 Query: 481 NHHSCYSGYCVKVGNEGF----CAKESMKSEKVGGGGPCVRDWQCYSRYCFNNGKLRYCA 648 C G CV GF CA + S+ G G CV QC F Sbjct: 507 QRGRCVDGRCVC--EHGFAGPDCADLACPSD-CHGRGRCVNG-QCVCHEGFT-------- 554 Query: 649 KDGKQFGEVGISGPCRKNKDCSSGRCVSMK--TKLQDGTESKVKVCQN 786 GK G+ G C C G+CV + T L G S C N Sbjct: 555 --GKDCGQRRCPGDCHGQGRCVDGQCVCHEGFTGLDCGQRSCPNDCSN 600
Score = 44.3 bits (103), Expect = 5e-04
Identities = 60/244 (24%), Positives = 90/244 (36%), Gaps = 11/244 (4%)
Frame = +1
Query: 31 CTEDKMCFSGVCVLLGNMQV--CAKDSKSSNLVGGGGACRRHVECFSGLCVKRDTGKVGF 204
C + C +GVCV C++++ H C G CV ++ GF
Sbjct: 225 CNDQGKCVNGVCVCFEGYSGVDCSRETCPVP------CSEEHGRCVDGRCVCQE----GF 274
Query: 205 CSKDSSVIKLIGGGGICKNNGQCYSRYCYKVDAETKICSKDSKFKGKVGGGGPCKNDFFC 384
+D + + C G+C C +C D F G+ G C D F
Sbjct: 275 AGEDCNEPLCLHN---CHGRGRCVENEC--------VC--DEGFTGEDCGELICPKDCFD 321
Query: 385 ASNFCHEINGIKYCVQESKQKIGYIGE--GAL-----CKNHHSCYSGYCVKVGNEGFCAK 543
ING YC + G+ GE G L C+ C G CV +EGF
Sbjct: 322 RGRC---INGTCYCDE------GFEGEDCGRLACPHGCRGRGRCEEGQCVC--DEGFAGA 370
Query: 544 ESMKSEKVGGGGPCVRDWQCYSRYCFNNGKLRYCAK--DGKQFGEVGISGPCRKNKDCSS 717
+ + C D C++R +G+ C +G+ GE+ G C + C +
Sbjct: 371 DCSERR-------CPSD--CHNRGRCLDGRCE-CDDGFEGEDCGELRCPGGCSGHGRCVN 420
Query: 718 GRCV 729
G+CV
Sbjct: 421 GQCV 424
Score = 32.0 bits (71), Expect = 2.4
Identities = 56/266 (21%), Positives = 82/266 (30%), Gaps = 29/266 (10%)
Frame = +1
Query: 19 GGGQCTEDK-MCFSGVCVLLGNMQVCAKDSKSSNLVGGGGACRRHVECFSGLCVKRDTGK 195
G G+C E++ +C G +C KD C C +G C +
Sbjct: 290 GRGRCVENECVCDEGFTGEDCGELICPKD------------CFDRGRCINGTCYCDE--- 334
Query: 196 VGFCSKDSSVIKLIGGGGICKNNGQCYSRYCY------KVDAETKICSKDSKFKGKVGGG 357
GF +D + G C+ G+C C D + C D
Sbjct: 335 -GFEGEDCGRLACPHG---CRGRGRCEEGQCVCDEGFAGADCSERRCPSD---------- 380
Query: 358 GPCKNDFFCASNFCHEINGIKYCVQESKQKIGYIGEGALCKNHHSCYSGYCV-KVGNEGF 534
C N C C +G + + G + C H C +G CV G G
Sbjct: 381 --CHNRGRCLDGRCECDDGFE------GEDCGELRCPGGCSGHGRCVNGQCVCDEGRTGE 432
Query: 535 CAKESMKSEKVGGGGPCVRDWQCYSRY---------------CFNNGKL--RYCAKD--- 654
+ G G CV+ +C + C +G+ C D
Sbjct: 433 DCSQLRCPNDCHGRGRCVQG-RCECEHGFQGYDCSEMSCPHDCHQHGRCVNGMCVCDDGY 491
Query: 655 -GKQFGEVGISGPCRKNKDCSSGRCV 729
G+ E+ G C + C GRCV
Sbjct: 492 TGEDCRELRCPGDCSQRGRCVDGRCV 517
>sp|P02876|AGI2_WHEAT Agglutinin isolectin 2 precursor (WGA2) (Isolectin D) Length = 213 Score = 43.9 bits (102), Expect = 6e-04 Identities = 49/169 (28%), Positives = 59/169 (34%), Gaps = 7/169 (4%) Frame = +1 Query: 241 GGGICKNNGQCYSRYCYKVDAETKICSKDSKFKGKVGGGGPCKNDFFCASNFCHEINGIK 420 GG C NN C S+Y + C +++ G GGPC+ D C S Sbjct: 78 GGATCPNN-HCCSQYGH--------CGFGAEYCGAGCQGGPCRADIKCGSQ--------- 119 Query: 421 YCVQESKQKIGYIGEGALCKNHHSCYSGYCVKVGNEGFCAKESMKSEKVGGG---GPCVR 591 G LC N+ C GFC + SE GGG G C Sbjct: 120 -------------SGGKLCPNNLCC--------SQWGFC---GLGSEFCGGGCQSGACST 155 Query: 592 DWQC----YSRYCFNNGKLRYCAKDGKQFGEVGISGPCRKNKDCSSGRC 726 D C R C NN YC ++G GI GP C SG C Sbjct: 156 DKPCGKDAGGRVCTNN----YCC---SKWGSCGI-GPGYCGAGCQSGGC 196
Score = 36.2 bits (82), Expect = 0.13
Identities = 38/139 (27%), Positives = 56/139 (40%), Gaps = 15/139 (10%)
Frame = +1
Query: 340 GKVGGGGPCKNDFFCASNFCHEINGIKYCVQE-------SKQKIGYIGEGALCKNHHSCY 498
G+ G C N+ C S + + G YC + + ++ G GA C N+H C
Sbjct: 31 GEQGSNMECPNNL-CCSQYGYCGMGGDYCGKGCQNGACWTSKRCGSQAGGATCPNNHCCS 89
Query: 499 S-GYCVKVGNEGFCAKESMKSEKVGGGGPCVRDWQCYS----RYCFNN---GKLRYCAKD 654
G+C GF A+ + GGPC D +C S + C NN + +C
Sbjct: 90 QYGHC------GFGAEYCGAGCQ---GGPCRADIKCGSQSGGKLCPNNLCCSQWGFCGLG 140
Query: 655 GKQFGEVGISGPCRKNKDC 711
+ G SG C +K C
Sbjct: 141 SEFCGGGCQSGACSTDKPC 159
>sp|P10969|AGI3_WHEAT Agglutinin isolectin 3 precursor (WGA3) Length = 186 Score = 43.9 bits (102), Expect = 6e-04 Identities = 48/163 (29%), Positives = 62/163 (38%), Gaps = 1/163 (0%) Frame = +1 Query: 241 GGGICKNNGQCYSRYCYKVDAETKICSKDSKFKGKVGGGGPCKNDFFCASNFCHEINGIK 420 GG C NN C S+Y + C +++ G GGPC+ D C S Sbjct: 51 GGKTCPNN-HCCSQYGH--------CGFGAEYCGAGCQGGPCRADIKCGSQ--------- 92 Query: 421 YCVQESKQKIGYIGEGALCKNHHSCYS-GYCVKVGNEGFCAKESMKSEKVGGGGPCVRDW 597 G LC N+ C GYC +G+E FC E ++ PC +D Sbjct: 93 -------------AGGKLCPNNLCCSQWGYC-GLGSE-FCG-EGCQNGACSTDKPCGKD- 135 Query: 598 QCYSRYCFNNGKLRYCAKDGKQFGEVGISGPCRKNKDCSSGRC 726 R C NN YC ++G GI GP C SG C Sbjct: 136 -AGGRVCTNN----YCC---SKWGSCGI-GPGYCGAGCQSGGC 169
>sp|P10040|CRB_DROME Crumbs protein precursor (95F) Length = 2146 Score = 42.7 bits (99), Expect = 0.001 Identities = 63/245 (25%), Positives = 86/245 (35%), Gaps = 20/245 (8%) Frame = +1 Query: 52 FSGVCVLLGNMQVCAKDSKSSNLVGGGGACRRHVECFSGLCVKRDTGKV---GFCSKDSS 222 FSG C+L G +S N+V G H G C D FC D Sbjct: 222 FSG-CLLDGPGLQFVNNSTVQNVVFG------HCPLTPGPCSDHDLFTRLPDNFCLNDPC 274 Query: 223 VIKLIGGGGICKNNGQCYSRYCYKVDAETKICSKDSKFKGKVGGGGPCKNDFFCASN--- 393 + G G C ++ + Y C K C KD+ G PC+N C N Sbjct: 275 M-----GHGTCSSSPEGYECRC-TARYSGKNCQKDN---GSPCAKNPCENGGSCLENSRG 325 Query: 394 ----FCHEINGIKYCVQESKQKIGYIGEGALCKNHHSCYSGYCVKVGNEGFCAKESMKSE 561 FC + ++C E + LC+ + +G CV +G G E K Sbjct: 326 DYQCFCDPNHSGQHCETE-------VNIHPLCQTNPCLNNGACVVIGGSGALTCECPKGY 378 Query: 562 KVGGGGPCVRDW-QCYSRYCFNNG----KLRYCAKDGKQFGEVGI-----SGPCRKNKDC 711 G C D +C S+ C NNG ++ + D G G C KN Sbjct: 379 ---AGARCEVDTDECASQPCQNNGSCIDRINGFSCDCSGTGYTGAFCQTNVDECDKNPCL 435 Query: 712 SSGRC 726 + GRC Sbjct: 436 NGGRC 440
Score = 34.3 bits (77), Expect = 0.49
Identities = 36/153 (23%), Positives = 54/153 (35%), Gaps = 11/153 (7%)
Frame = +1
Query: 100 DSKSSNLVGGGGACRRHVECFSGLCVKRDTGKVGFCSKDSSVIKLIGGGGICKNNGQCYS 279
D SN G C + F+ C+ TG++ D V G C N GQC
Sbjct: 725 DECGSNPCSNGSTCIDRINNFTCNCIPGMTGRICDIDIDDCV------GDPCLNGGQCID 778
Query: 280 RY------CYKVDAETKIC--SKDSKFKGKVGGGGPCKN---DFFCASNFCHEINGIKYC 426
+ C E + C + D G C + D+FC CH K C
Sbjct: 779 QLGGFRCDCSGTGYEGENCELNIDECLSNPCTNGAKCLDRVKDYFCD---CHNGYKGKNC 835
Query: 427 VQESKQKIGYIGEGALCKNHHSCYSGYCVKVGN 525
Q+ + C+++ Y+G C++ N
Sbjct: 836 EQDINE----------CESNPCQYNGNCLERSN 858
Score = 33.1 bits (74), Expect = 1.1
Identities = 61/280 (21%), Positives = 91/280 (32%), Gaps = 53/280 (18%)
Frame = +1
Query: 31 CTEDKMCFSGVCVLLGNM-------QVCAKDSKSSNLVGGGGACRRHVECFSGLCVKRDT 189
C E +C C+ N Q C + +L G C+ C + T
Sbjct: 589 CQEPMVCVQNQCLCPENKVCNQCATQPCQNGGECVDLPNGDYECK---------CTRGWT 639
Query: 190 GKVGFCSKDSSVIKL---IGGGGICKNNGQCYSRYC------------------------ 288
G+ C D L I G GICKN Y YC
Sbjct: 640 GRT--CGNDVDECTLHPKICGNGICKNEKGSYKCYCTPGFTGVHCDSDVDECLSFPCLNG 697
Query: 289 ----YKVDAETKIC-----SKDSKFKGKVGGGGPCKNDFFC---ASNF-CHEINGI--KY 423
K++A +C ++ + G PC N C +NF C+ I G+ +
Sbjct: 698 ATCHNKINAYECVCQPGYEGENCEVDIDECGSNPCSNGSTCIDRINNFTCNCIPGMTGRI 757
Query: 424 CVQESKQKIG-YIGEGALCKNHHSCYSGYCVKVGNEGFCAK---ESMKSEKVGGGGPCVR 591
C + +G G C + + C G EG + + S G C+
Sbjct: 758 CDIDIDDCVGDPCLNGGQCIDQLGGFRCDCSGTGYEGENCELNIDECLSNPCTNGAKCLD 817
Query: 592 DWQCYSRYCFNNGKLRYCAKDGKQFGEVGISGPCRKNKDC 711
+ Y C N K + C +D + S PC+ N +C
Sbjct: 818 RVKDYFCDCHNGYKGKNCEQDINECE----SNPCQYNGNC 853
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 112,855,461
Number of Sequences: 369166
Number of extensions: 2835641
Number of successful extensions: 8118
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7014
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7995
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 9126484700
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail