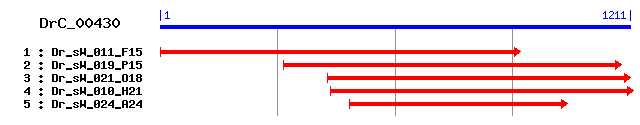
DrC_00430
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00430 (1211 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O75095|EGFL3_HUMAN Multiple EGF-like-domain protein 3 pr... 38 0.051 sp|O88281|EGFL3_RAT Multiple EGF-like-domain protein 3 prec... 35 0.33 sp|Q8VHS2|CRUM1_MOUSE Crumbs protein homolog 1 precursor 34 0.97 sp|Q3LI77|KR134_HUMAN Keratin-associated protein 13-4 33 1.6 sp|Q5K651|SAMD9_HUMAN Sterile alpha motif domain containing... 32 2.8 sp|Q80V70|EGFL3_MOUSE Multiple EGF-like-domain protein 3 32 3.7 sp|O18735|ERBB2_CANFA Receptor tyrosine-protein kinase erbB... 31 8.2 sp|P46213|SYI_THEMA Isoleucyl-tRNA synthetase (Isoleucine--... 31 8.2 sp|Q60553|ERBB2_MESAU Receptor tyrosine-protein kinase erbB... 31 8.2
>sp|O75095|EGFL3_HUMAN Multiple EGF-like-domain protein 3 precursor (Multiple epidermal growth factor-like domains 6) Length = 1229 Score = 38.1 bits (87), Expect = 0.051 Identities = 25/84 (29%), Positives = 35/84 (41%), Gaps = 1/84 (1%) Frame = +2 Query: 488 GGNCEHPSVNFTFG-GVYQVCDGPAGPSYSQVNPLTGSNSCPKGFSSVKLHSGLIRCDTS 664 G +CE +G G ++C PA ++ +P TG+ C GF RC Sbjct: 661 GEDCEADCPEGRWGLGCQEIC--PACQHAARCDPETGACLCLPGFVGS-------RCQDV 711 Query: 665 CSGWWFWRHCDTRCSYFTTYWCYP 736 C W+ C TRCS C+P Sbjct: 712 CPAGWYGPSCQTRCSCANDGHCHP 735
Score = 32.3 bits (72), Expect = 2.8
Identities = 25/82 (30%), Positives = 32/82 (39%), Gaps = 7/82 (8%)
Frame = +2
Query: 581 NPLTGSNSCPKGFSSVKLHSGLIRCDTSCSGWWFWRHCDTRCSYFTTYWCYPNAVGENHN 760
+P TG CP G++ K C + C WF C RCS C P A +
Sbjct: 907 DPHTGRCLCPAGWTGDK-------CQSPCLRGWFGEACAQRCS------CPPGAACHHVT 953
Query: 761 -------GYQFGGIKQGNCPIG 805
G+ G +QG CP G
Sbjct: 954 GACRCPPGFTGSGCEQG-CPPG 974
>sp|O88281|EGFL3_RAT Multiple EGF-like-domain protein 3 precursor (Multiple epidermal growth factor-like domains 6) Length = 1574 Score = 35.4 bits (80), Expect = 0.33 Identities = 21/69 (30%), Positives = 28/69 (40%) Frame = +2 Query: 530 GVYQVCDGPAGPSYSQVNPLTGSNSCPKGFSSVKLHSGLIRCDTSCSGWWFWRHCDTRCS 709 G ++C PA + NP TG+ C GF RC +CS W+ C RC+ Sbjct: 785 GCQEIC--PACEHGASCNPETGTCLCLPGFVGS-------RCQDTCSAGWYGTGCQIRCA 835 Query: 710 YFTTYWCYP 736 C P Sbjct: 836 CANDGHCDP 844
Score = 31.6 bits (70), Expect = 4.8
Identities = 24/83 (28%), Positives = 32/83 (38%), Gaps = 1/83 (1%)
Frame = +2
Query: 488 GGNCEHPSVNFTFG-GVYQVCDGPAGPSYSQVNPLTGSNSCPKGFSSVKLHSGLIRCDTS 664
G C+ V+ TFG + C G S V TG+ CP G+ C+ +
Sbjct: 1116 GDKCQSSCVSGTFGVHCEEHCACRKGASCHHV---TGACFCPPGWRGP-------HCEQA 1165
Query: 665 CSGWWFWRHCDTRCSYFTTYWCY 733
C WF C RC T C+
Sbjct: 1166 CPRGWFGEACAQRCLCPTNASCH 1188
Score = 31.2 bits (69), Expect = 6.3
Identities = 26/79 (32%), Positives = 29/79 (36%), Gaps = 4/79 (5%)
Frame = +2
Query: 581 NPLTGSNSCPKGFSSVKLHSGLIRCDTSCSGWWFWRHCDTRCSYFTTYWCYPNAVGENHN 760
NP GS SC GF RC C +F C RC+ C P GE
Sbjct: 669 NPKDGSCSCKAGFQGE-------RCQAECESGFFGPGCRHRCTCQPGVACDP-VSGECRT 720
Query: 761 ----GYQFGGIKQGNCPIG 805
GYQ Q CP+G
Sbjct: 721 QCPPGYQGEDCGQ-ECPVG 738
Score = 30.8 bits (68), Expect = 8.2
Identities = 32/109 (29%), Positives = 40/109 (36%), Gaps = 3/109 (2%)
Frame = +2
Query: 488 GGNCEHPSVNFTFG-GVYQVCDGPAGPSYSQVNPLTGSNSCPKGFSSVKLHSGLIRCDTS 664
G +CE FG Q C P S V TG CP GF+ + C+ +
Sbjct: 1159 GPHCEQACPRGWFGEACAQRCLCPTNASCHHV---TGECRCPPGFTG-------LSCEQA 1208
Query: 665 CSGWWFWRHCDTRCSYFTTYW-CYP-NAVGENHNGYQFGGIKQGNCPIG 805
C F + C+ C W C P + V GY G Q CP G
Sbjct: 1209 CQPGTFGKDCEHLCQCPGETWACDPASGVCTCAAGYHGTGCLQ-RCPSG 1256
>sp|Q8VHS2|CRUM1_MOUSE Crumbs protein homolog 1 precursor Length = 1405 Score = 33.9 bits (76), Expect = 0.97 Identities = 55/226 (24%), Positives = 83/226 (36%), Gaps = 16/226 (7%) Frame = +2 Query: 434 KGCLDFTSPNFNSDANVDGGNCEHPSV-NFTFGGVYQVCDGPAGPSYSQVNPLTGSNSCP 610 K C D P F+S + P NF +C P P YS +N T +NSC Sbjct: 64 KDCEDLKDPCFSSPCQGIATCVKIPGEGNF-------LCQCP--PGYSGLNCETATNSCG 114 Query: 611 KGFSSVKLHSGLIRCDTS-----CSGWWFWRHCDTRCSYFTTYWCYPNAVGENH-NGYQF 772 ++ H G R D C + R C+T + + C+ A+ ++ NGY Sbjct: 115 ---GNLCQHGGTCRKDPEHPVCICPPGYAGRFCETDHNECASSPCHNGAMCQDGINGYSC 171 Query: 773 GGIKQGNCPIGYISLKVGLSVEICVTIDNDPNNPFAIKFGGLFSC-------SVGNPLAK 931 C GY L V+ CV+ D N + G ++C V L Sbjct: 172 ------FCVPGYQGRHCDLEVDECVS-DPCKNEAVCLNEIGRYTCVCPQEFSGVNCELEI 224 Query: 932 EFVKGKPKLSSSKMMDLV-YWQKTCAPGYI-SHIASIEQGCQISYC 1063 + + +P L + D + CAPG++ H C+ C Sbjct: 225 DECRSQPCLHGATCQDAPGGYSCDCAPGFLGEHCELSVNECESQPC 270
>sp|Q3LI77|KR134_HUMAN Keratin-associated protein 13-4 Length = 160 Score = 33.1 bits (74), Expect = 1.6 Identities = 27/96 (28%), Positives = 39/96 (40%) Frame = +2 Query: 539 QVCDGPAGPSYSQVNPLTGSNSCPKGFSSVKLHSGLIRCDTSCSGWWFWRHCDTRCSYFT 718 + C PA S P T CP C T+CSG +R R + Sbjct: 53 KTCWEPASCQKSCYRPRTSILCCP--------------CQTTCSGSLGFRSSSCRSQGYG 98 Query: 719 TYWCYPNAVGENHNGYQFGGIKQGNCPIGYISLKVG 826 + CY ++G +G++F +K G C G+ SL G Sbjct: 99 SRCCY--SLGNGSSGFRF--LKYGGC--GFPSLSYG 128
>sp|Q5K651|SAMD9_HUMAN Sterile alpha motif domain containing protein 9 Length = 1589 Score = 32.3 bits (72), Expect = 2.8 Identities = 21/78 (26%), Positives = 40/78 (51%), Gaps = 2/78 (2%) Frame = +2 Query: 2 HVIRSIDAGAILSKTDALKSTYVKKLNS--DTTTISVSATASFLNLFGMSAKYSTQSSQT 175 +V+R+I G + +A +++ LNS TTIS+S FL + A + T+ + Sbjct: 899 NVVRNILKGQNIFTKEAKLFSFLALLNSYVPDTTISLSQCEKFLGIGNKKAFWGTEKFED 958 Query: 176 QIDEYNTNVARTHIDTVG 229 ++ Y+T + +T + G Sbjct: 959 KMGTYSTILIKTEVIECG 976
>sp|Q80V70|EGFL3_MOUSE Multiple EGF-like-domain protein 3 Length = 656 Score = 32.0 bits (71), Expect = 3.7 Identities = 31/125 (24%), Positives = 46/125 (36%), Gaps = 1/125 (0%) Frame = +2 Query: 437 GCLDFTSPNFNSDANVDGGNCEHPSVNFTFG-GVYQVCDGPAGPSYSQVNPLTGSNSCPK 613 G D + + A G C+ P V+ FG + C G + V TG+ CP Sbjct: 181 GTCDRLTGHCRCPAGWTGDKCQSPCVSGMFGVHCEEHCACRKGATCHHV---TGACLCPP 237 Query: 614 GFSSVKLHSGLIRCDTSCSGWWFWRHCDTRCSYFTTYWCYPNAVGENHNGYQFGGIKQGN 793 G+ C+ +C WF C RC C P A + +G + + Sbjct: 238 GWRGS-------HCEQACPRGWFGEACAQRCH------CPPGASCHHVSG-------ECH 277 Query: 794 CPIGY 808 CP G+ Sbjct: 278 CPPGF 282
>sp|O18735|ERBB2_CANFA Receptor tyrosine-protein kinase erbB-2 precursor (p185erbB2) (C-erbB-2) Length = 1259 Score = 30.8 bits (68), Expect = 8.2 Identities = 31/119 (26%), Positives = 42/119 (35%), Gaps = 1/119 (0%) Frame = +2 Query: 491 GNCEHPSVNFTFGGVYQVCDGPAGPSYSQVNPLTGSNSCPKGFSSVKLHSGLIRCDTSCS 670 G+C+ + GG + C GP C G + K HS + C Sbjct: 210 GDCQSLTRTVCAGGCAR-CKGPQPTDCCH-------EQCAAGCTGPK-HSDCLACLHFNH 260 Query: 671 GWWFWRHCDTRCSYFT-TYWCYPNAVGENHNGYQFGGIKQGNCPIGYISLKVGLSVEIC 844 HC +Y T T+ PN G Y FG +CP Y+S VG +C Sbjct: 261 SGICELHCPALVTYNTDTFESMPNPEGR----YTFGASCVTSCPYNYLSTDVGSCTLVC 315
>sp|P46213|SYI_THEMA Isoleucyl-tRNA synthetase (Isoleucine--tRNA ligase) (IleRS) Length = 919 Score = 30.8 bits (68), Expect = 8.2 Identities = 18/61 (29%), Positives = 30/61 (49%), Gaps = 3/61 (4%) Frame = +2 Query: 257 LKEWEMSIMDNLVAIDRRGEPLYYAITPHALPDLDDGTVH---KINVLLRKAIERYYVRN 427 L+EWE + N V R+G+PL+ H P +G +H +N +L+ + +Y Sbjct: 28 LEEWEKMDLYNYVLEQRKGKPLFVL---HDGPPYANGHIHIGTALNKILKDIVVKYKTMR 84 Query: 428 G 430 G Sbjct: 85 G 85
>sp|Q60553|ERBB2_MESAU Receptor tyrosine-protein kinase erbB-2 precursor (p185erbB2) (C-erbB-2) (NEU proto-oncogene) Length = 1254 Score = 30.8 bits (68), Expect = 8.2 Identities = 18/53 (33%), Positives = 23/53 (43%), Gaps = 1/53 (1%) Frame = +2 Query: 689 HCDTRCSYFT-TYWCYPNAVGENHNGYQFGGIKQGNCPIGYISLKVGLSVEIC 844 HC +Y T T+ PN G Y FG CP Y+S +VG +C Sbjct: 267 HCPALVTYNTDTFESMPNPEGR----YTFGASCVTTCPYNYLSTEVGSCTLVC 315
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 143,229,384
Number of Sequences: 369166
Number of extensions: 3247017
Number of successful extensions: 7592
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 7178
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7584
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 13769178250
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail